Transcriptional control of mycobacterial DNA damage response by sigma adaptation
A recent study by the Weber-Ban and Ban groups (IMBB) published in Science Advances uncovers that the master regulator of the DNA damage response in mycobacteria, PafBC, leverages a unique mechanism of transcriptional activation to allow promoter recognition at promoters lacking the canonical -35 motif.
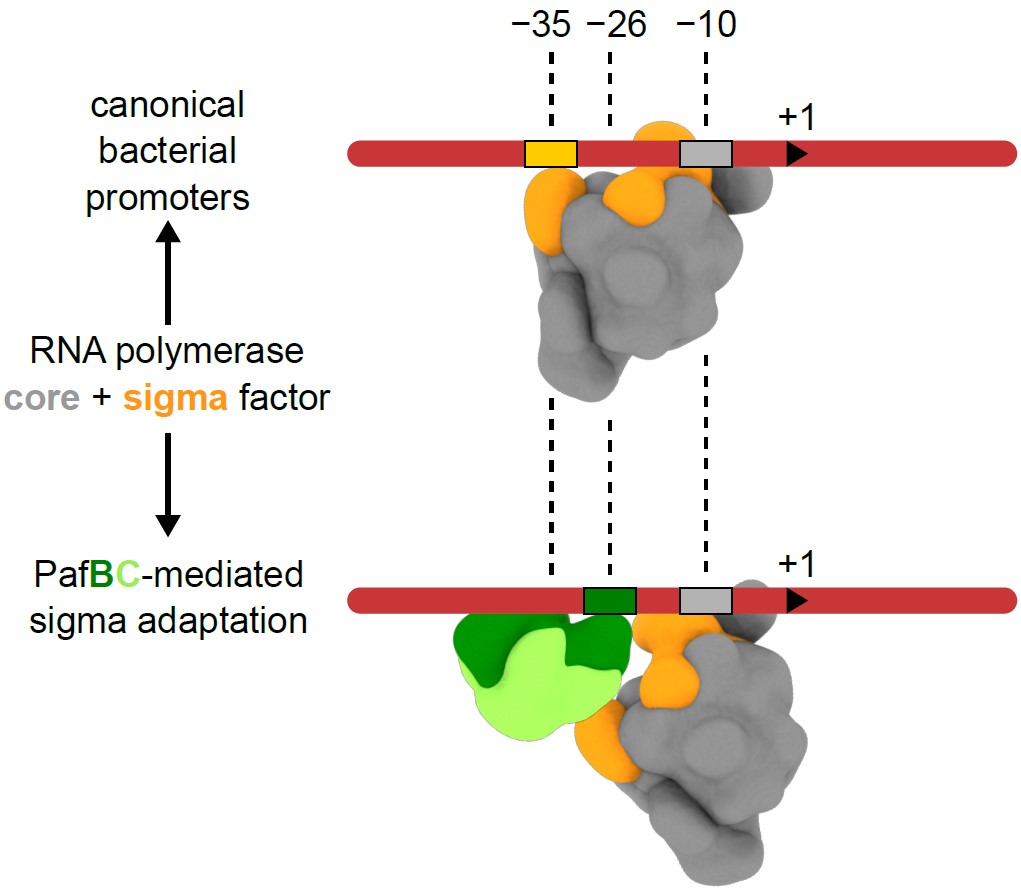
At the transcriptional level, mycobacteria, such as the human pathogen M. tuberculosis, respond to DNA damage via two intertwined regulatory pathways. One pathway constitutes the canonical SOS response, which is regulated by RecA and repressor LexA. The other, recently discovered pathway under control of regulator PafBC controls a large number of genes by transcription activation, including recA. While the derepression mechanism of the SOS response is well understood, PafBC’s mechanism of transcription activation remained unknown.
Researchers at IMBB discovered a unique mechanism of promoter recognition employed by PafBC. In bacteria, promoter recognition usually involves two canonical sequence motifs located at positions -35 and -10 from the transcription start site. However, PafBC-dependent promoters can be considered “hybrid”, since they include the canonical -10 region but lack the -35 motif and instead feature a PafBC-specific sequence motif at position -26. Using cryo-electron microscopy and biochemical experiments, the researchers visualized a PafBC-containing transcription initiation complex. They show that PafBC functions as an adaptor by inserting between the sigma subunit and the DNA so that the RNA polymerase holoenzyme recognizes a hybrid -26/-10 promoter, hence this mechanism is referred to as ‘sigma adaptation’.
Genome-wide studies found that many promoters in mycobacteria lack a canonical -35 motif. The mechanism of PafBC-dependent ‘sigma adaptation’ could therefore represent an example of a widespread, alternative mode of bacterial promoter recognition employed also by other transcription factors.
Link to the paper in external page Science Advances