Structural basis of adenylyl cyclase 9 activation
The recent Nature Communications paper by the groups of Volodymyr Korkhov (IMBB, ETHZ & PSI), in collaboration with Plückthun (UZH), Dessauer (UTHealth Houston) and Bondar (FZ Jülich & University of Bucharest) groups describes the conformational changes in membrane adenylyl cyclase AC9 induced by different activators.
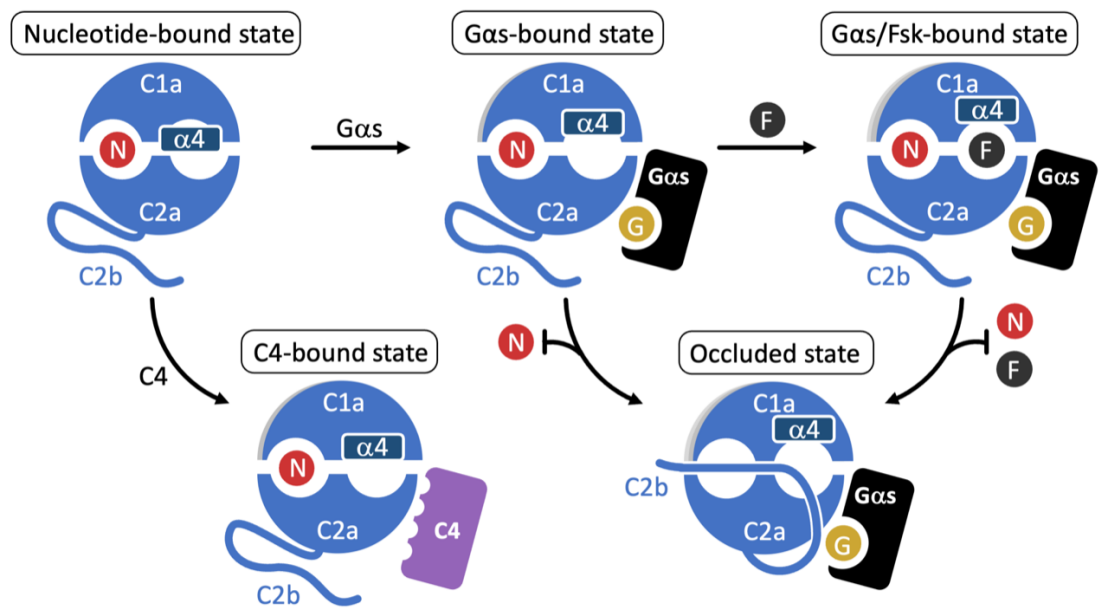
Adenylyl cyclase 9 (AC9) is a membrane-bound enzyme that converts ATP into cAMP. The enzyme is weakly activated by a small molecule activator forskolin, fully activated by the G protein Gαs subunit and is autoinhibited by the AC9 C-terminus. Although the structural studies of the AC9-Gαs complex provided the framework for understanding AC9 autoinhibition, the conformational changes that AC9 undergoes in response to activator binding remained poorly understood.
In collaboration with the groups of Andreas Plückthun (University of Zurich), Carmen Dessauer (UTHealth Houston) and Ana-Nicoleta Bondar (Forschungszentrum Jülich & University of Bucharest), the team of Volodymyr Korkhov (Institute of Molecular Biology and Biophysics, ETH Zurich & Paul Scherrer Institute) determined the cryo-EM structures of AC9 in several distinct states: (i) AC9 bound to a nucleotide inhibitor MANT-GTP, (ii) bound to an artificial activator (DARPin C4) and MANT-GTP, (iii) bound to DARPin C4 and a nucleotide analogue ATPαS, (iv) bound to Gαs and MANT-GTP. The artificial activator DARPin C4 partially activates AC9 by binding at a site that overlaps with the Gαs binding site. Together with the previously observed occluded and forskolin-bound conformations, comparisons of AC9 in these four new states revealed the structural rearrangements essential for AC9 activation.
Link to the paper in external page Nature Communications