The cassava genome revisited to accelerate breeding efforts
A recent GigaScience paper by the Gruissem group (IMPB) in collaboration with the Functional Genomics Center Zurich uses cutting-edge long-range DNA sequencing technology to generate the most accurate assembly of the highly heterozygous cassava genome, which accelerates breeding of this important crop.

Cassava is one of the five most important crops in the world and feeds about one billion people, mostly in tropical countries. The starchy storage root is eaten boiled, fried or roasted, and leaves are often used as a vegetable. Cassava flour is gluten-free and popular for baking cakes and as starchy balls in bubble teas, especially in Southeast Asia. Cassava is vegetatively propagated by farmers and considered a food security crop because it is tolerant to drought, and farmers can leave the starchy storage root in the ground until needed. However, breeding new cassava varieties that are resistant to diseases, especially virus infections that often devastate production in Sub-Saharan Africa, India and more recently in Southeast Asia as well, is difficult because the cassava genome is highly heterozygous. An accurate assembly of the cassava genome would therefore be helpful for breeders to guide their efforts.

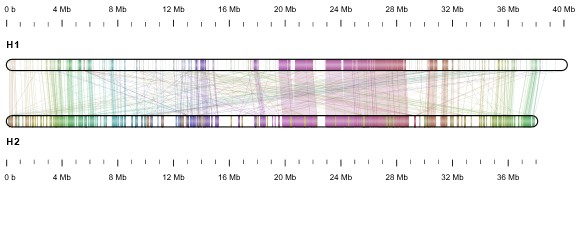
The team led by Weihong Qi and Wilhelm Gruissem used Pacific Biosciences High Fidelity (HiFi) sequencing in combination with innovative assemblers and Hi-C phasing technology to produce a nearly complete (99%) heterozygous diploid cassava genome at chromosome scale and haplotype resolution. Although other cassava genomes had been reported earlier (including from the Gruissem lab), because of their complex nature they lacked the resolution and accuracy that has now been achieved. The genome assembly revealed about 35,000 phased gene pairs and extensive chromosome rearrangements with large structural variations caused by retrotransposons. Many of the genes have differences in the DNA sequences of their two copies (alleles) and often only one of the two alleles is expressed in different tissues. The team has used the reference-quality chromosome assemblies to build a pan-genome that represents the genetic diversity of cassava, which is important for reference-guided functional genomics analysis and breeding strategies.
Link to the paper in external page GigaScience