Environmental genome reconstruction greatly improves microbial taxon profiling tool mOTUs
A collaborative effort, published in Microbiome, between the Sunagawa lab and the Zeller team (EMBL, Heidelberg) resulted in mOTUs3: a taxonomic profiling tool to detect and quantify more than 33,500 prokaryotic species, most of which represent uncultivated microbes from >20 different environments.
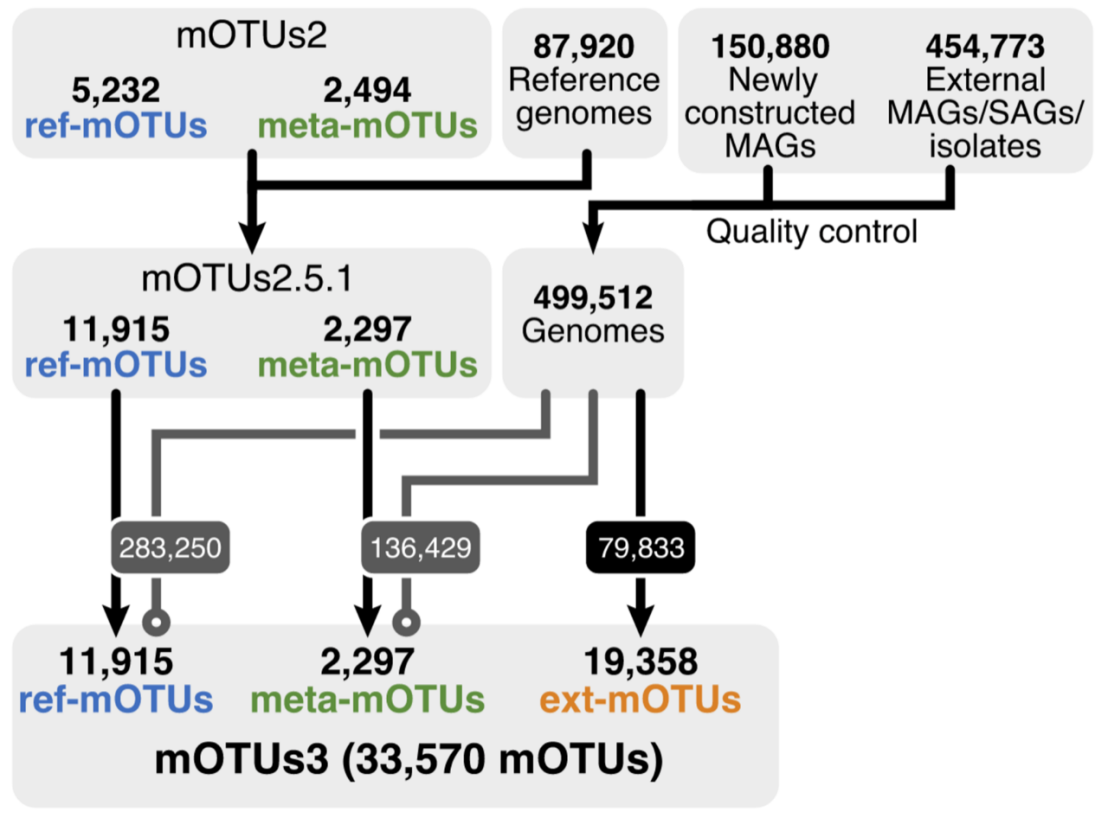
Taxonomic profiling is a fundamental task in microbiome research that aims to detect and quantify the relative abundance of microorganisms in biological samples. However, a substantial fraction of the microbial species present in many environments still remains unaccounted for during taxonomic profiling of shotgun metagenomic data as we lack sequenced reference genomes to compare against. To address this issue, we present mOTUs3, a software tool which profiles metagenomes for 33.5k species-level operational taxonomic units, two thirds of which are currently not represented by reference genomes. To facilitate reference genome-independent profiling, we collected a set of phylogenetic marker gene sequences from metagenome-assembled and single cell genomes from 23 environments, including soil, freshwater systems, and the gastrointestinal tract of ruminants and other animals, for which only a few whole microbial genomes exist. We illustrate how mOTUs3 greatly increases the microbial diversity captured across these environments and the number of differentially abundant species, including novel ones, in cross-sectional studies.
Link to the paper in external page Microbiome