Millions of reconstructed genomes from diverse microbiomes at global scale
A recent “Nucleic Acids Research” paper by the Sunagawa group (IMB) showcases the new mOTUs online database (mOTUs-db), a resource which allows to select and download genomes from host- associated and environmental microbiomes, representing the largest prokaryotic genome collection to date.
Microbes are the most numerically dominant and phylogenetically diverse organisms on Earth. They drive biogeochemical cycling of energy, biomass and nutrients and are crucial to the health of their macroscopic hosts such as humans, animals and plants.
A fundamental task in studying microbial communities is determining which and how many microbes are present (i.e., taxonomic profiling). Comparison of taxonomic profiles has been leveraged, for example, to develop biomarkers for diseases such as colorectal cancer or to assess the likelihood of response to cancer immunotherapy. Many detected microbes, however, lack a genomic representative, which prevents the assessment of their genomic context and gaining insight into their potential function.
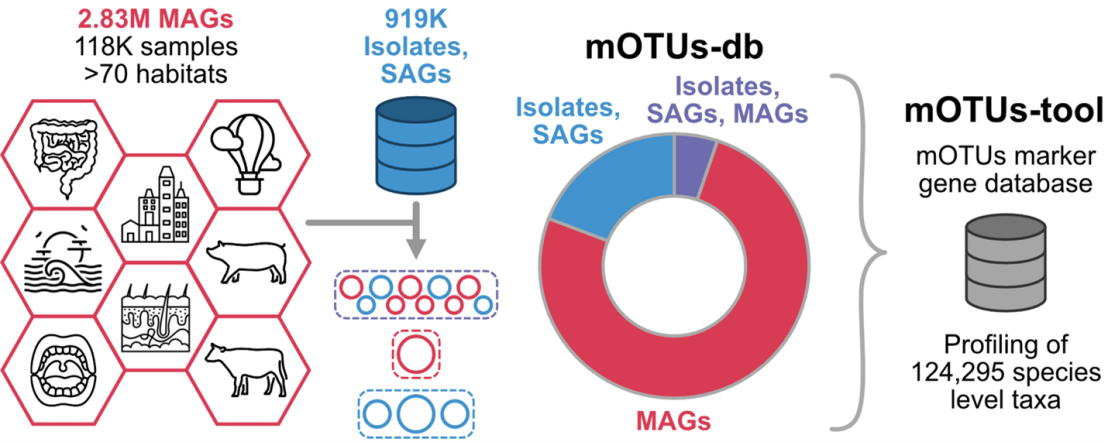
To increase the scope of available genome-resolved microbial diversity, the Sunagawa group (IMB) has systematically reconstructed 2.83M metagenome-assembled genomes (MAGs) from >117K environmental samples covering >70 distinct habitats, e.g. humans, farm animals, lakes, rivers, and oceans. These 2.83M MAGs were supplemented with 919,090 genomes from public genome databases and clustered into 124,295 species-level taxonomic units, with over 50% species unique to the database. The database interfaces with the mOTUs taxonomic profiling tool, thus enabling users to interactively explore and download the genomes of microorganisms detected within their sample.
Link to the paper in external page Nucleic Acids Research.