Molecular features of substrate recognition in the mycobacterial Bpa-proteasome complex
A new article by the Weber-Ban group, in collaboration with the Glockshuber group and the cryoEM knowledge hub, reveals how the mycobacterial proteasome activator Bpa binds substrates via key ring-lining residues, shedding light on protein degradation in M. tuberculosis.
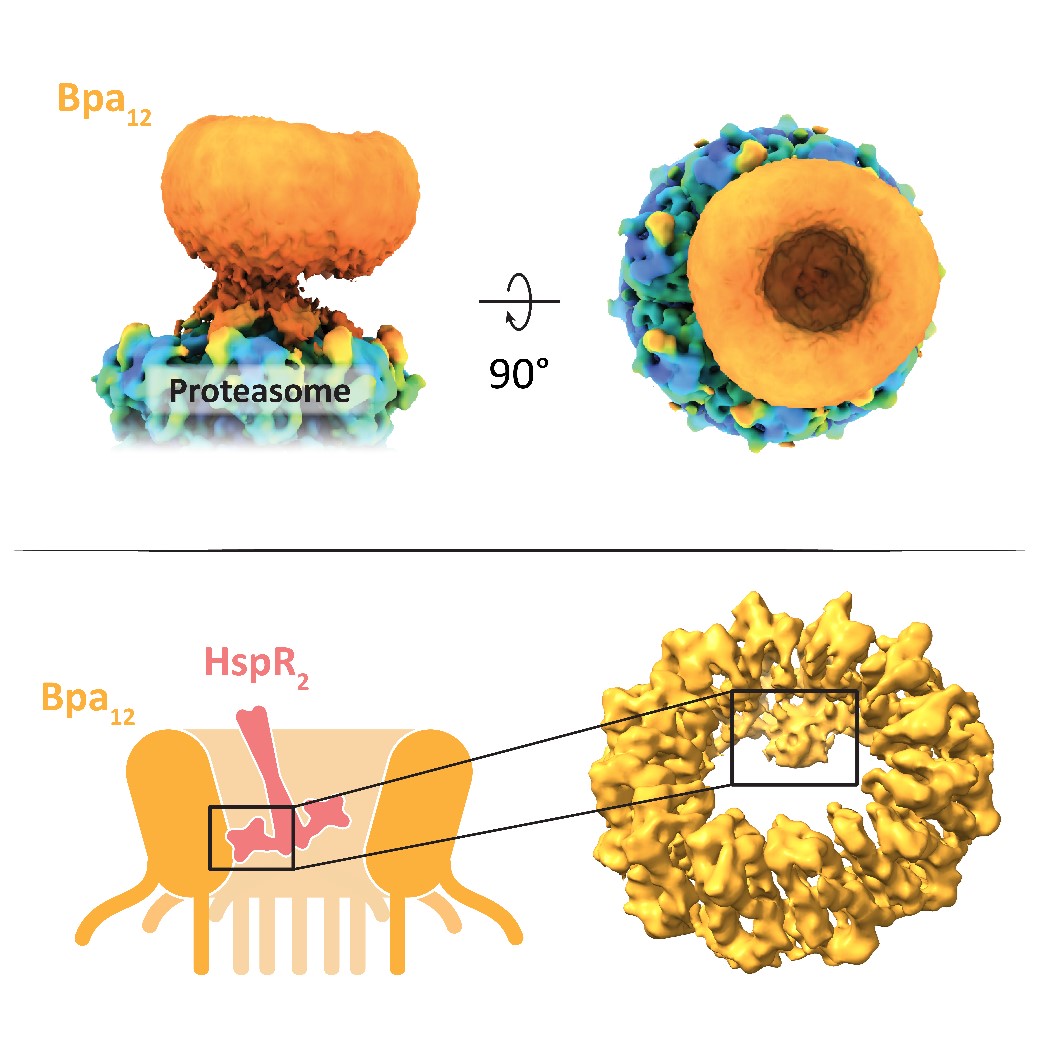
Compartmentalizing proteases enable the regulated degradation of proteins. Over the course of evolution, mycobacteria have acquired a eukaryotic-like proteasome, providing them with a selective advantage under stress conditions, such as those encountered by Mycobacterium tuberculosis during host infection. The bacterial proteasome relies on activator complexes, like the bacterial proteasome activator (Bpa), to select its substrates. Bpa functions independently of ATP and does not recognize any known molecular degradation tags on proteins. The Weber-Ban group has previously identified the molecular and structural features required for substrates to be degraded by the Bpa-proteasome complex. However, the precise mechanism by which proteins are recognized by the activator has remained unclear.
Using cryo-electron microscopy, the authors obtained a 3D reconstruction of the Bpa-proteasome complex, revealing that Bpa interacts asymmetrically with the proteasome, adopting a “hinged lid” conformation. Crosslinking, combined with cryo-EM and mass spectrometry, allowed the authors to pinpoint specific residues in Bpa that are critical for the degradation of its natural substrate HspR. Together, these findings raise intriguing questions about the physiological relevance of the hinged lid conformation and the mechanism of substrate engagement and translocation. Gaining insight into these processes advances our understanding of bacterial survival strategies and may contribute to the development of novel antimicrobial therapies.
Link to the paper in external page Nature Communications.