Predicting input signals of transcription factors in E. coli
A recent "Molecular Systems Biology" paper by the Sauer group (IMSB) systematically predicted intracellular metabolite input signals for 41 E. coli transcription factors.
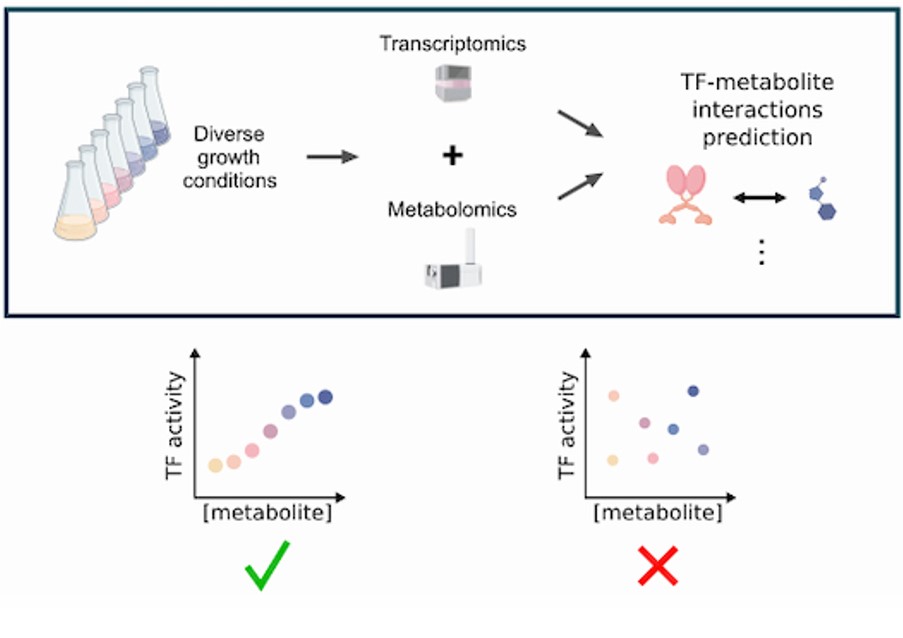
To effectively elicit gene expression responses, it is essential that information from the extracellular environment or the cellular state is transmitted to transcription factors (TFs). In bacteria, the predominant mechanism for this signaling is through allosteric binding of intracellular metabolites. In this study, we developed a systematic workflow for identifying the input signals of bacterial TFs by utilizing metabolomics and transcriptomics data. We inferred the activity of 173 TFs from existing transcriptomics data and determined the abundance of 279 metabolites across 40 matched experimental conditions in E. coli. By correlating TF activities with metabolite abundances, we successfully confirmed previously known TF–metabolite interactions and predicted novel effectors metabolites for 41 TFs. Our established regulatory network includes 76 novel interactions.
Link to the paper in external page "Molecular Systems Biology".