Extensive regulation of enzyme activity by phosphorylation in E. coli
A recent "Nature Communications" paper by the Sauer group (IMSB) demonstrates surprisingly frequent metabolic phenotypes of point mutations in phosphosites of enzymes, suggesting protein phosphorylation is an underappreciated major regulation process in bacterial metabolism.
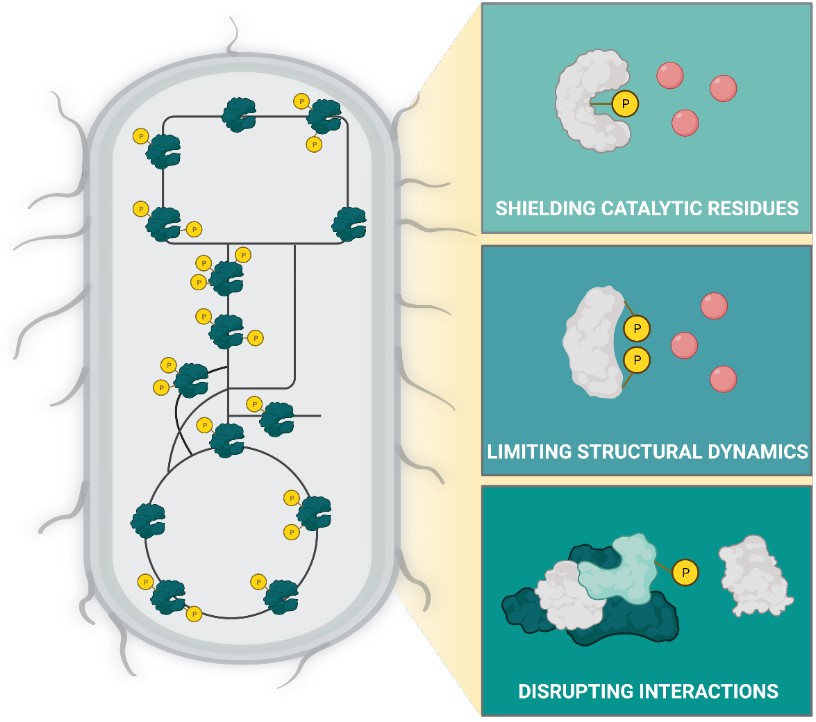
Protein phosphorylation is an essential and frequent post-translation modification in eukaryotes, but historically has been considered less prevalent in bacteria because fewer proteins were found to be phosphorylated and most proteins were modified to a lower degree. Recent proteomics studies greatly expanded the phosphoproteome of E. coli to more than 2000 phosphosites, yet mechanisms of action were proposed for only six of these phosphosites and fitness effects are known for 38 of them.
By systematically characterizing functional relevance of protein phosphorylation in E. coli metabolism, the Sauer group found 44 of the 52 mutated phosphosites to be functional based on growth phenotypes and intracellular metabolome profiles. By effectively doubling the number of known functional phosphosites and finding that nearly all investigated sites exhibit metabolic phenotypes, they provide evidence that protein phosphorylation is a major regulation process in bacterial metabolism. Combining in vitro and in vivo experiments, they demonstrate how single phosphosites modulate enzymatic activity and regulate metabolic fluxes in glycolysis, methylglyoxal bypass, acetate metabolism and the split between pentose phosphate and Entner-Doudoroff pathways through mechanisms that include shielding the substrate binding site, limiting structural dynamics, and disrupting interactions relevant for activity in vivo.
Link to the paper in external page Nature Communications