NabFab: A nanobody-binding Fab for fiducial-assisted cryo-EM studies of membrane proteins
Small integral membrane proteins comprise a large portion of drug targets in humans. These proteins are often resistant to structure determination. A recent "PNAS" by the Locher group (IMBB) in collaboration with the Kossiakoff Group (University of Chicago) describes a widely applicable solution.
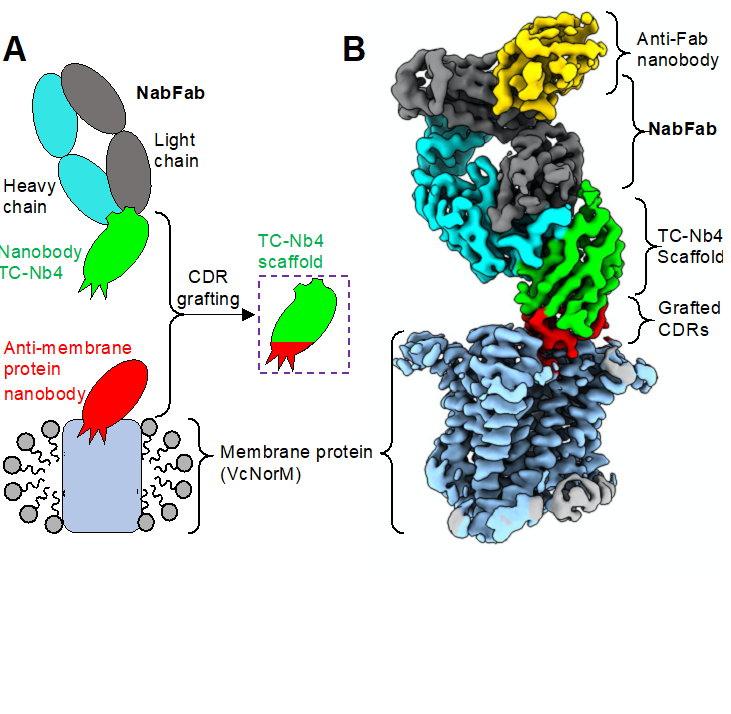
While single-particle cryo-EM has become a powerful tool for the structure determination of many proteins that otherwise are resistant to crystallization, small integral membrane proteins remain particularly challenging targets. This is because they often lack distinct features that allow for useful particle alignment, and most of the protein is masked by a surrounding detergent micelle or nanodisc bilayer. Antigen-binding fragments (Fabs) binding to small membrane proteins have become a handy tool to overcome this "size limit" in cryo-EM structure determination. Such Fabs can act as fiducial marks in single-particle analysis by adding a distinct structural feature to their targets. However, Fab generation is often challenging. A smaller class of antibodies, called nanobodies, have been widely used for their properties in locking membrane proteins in distinct conformational states and as crystallization chaperones.
Bloch J.S. and Mukherjee S. et al. describe the development of a Fab (NabFab) that rigidly binds to the scaffold of nanobodies. The added mass and shape of the NabFab facilitates cryo-EM studies, as they demonstrate with the structure determination of two ˜50 kDa sized membrane proteins. Furthermore, a simple grafting strategy ensures the universal binding of NabFab to nanobodies of different origins. Thus, the general applicability of NabFab might enable the structure determination of many existing membrane-nanobody protein complexes, including SLCs and GPCRs.
Link to the paper in external page PNAS