Structures of Pup in complex with depupylase Dop reveal the mechanism of catalytic Pi formation
Mycobacteria mark proteins by pupylation, a process akin to ubiquitination. A pair of related enzymes catalyze attachment and removal of Pup. A Nature Communications article by the Weber-Ban lab reveals how depupylase Dop generates the catalytic phosphate that enables Dop to cleave the bond between Pup and targets.
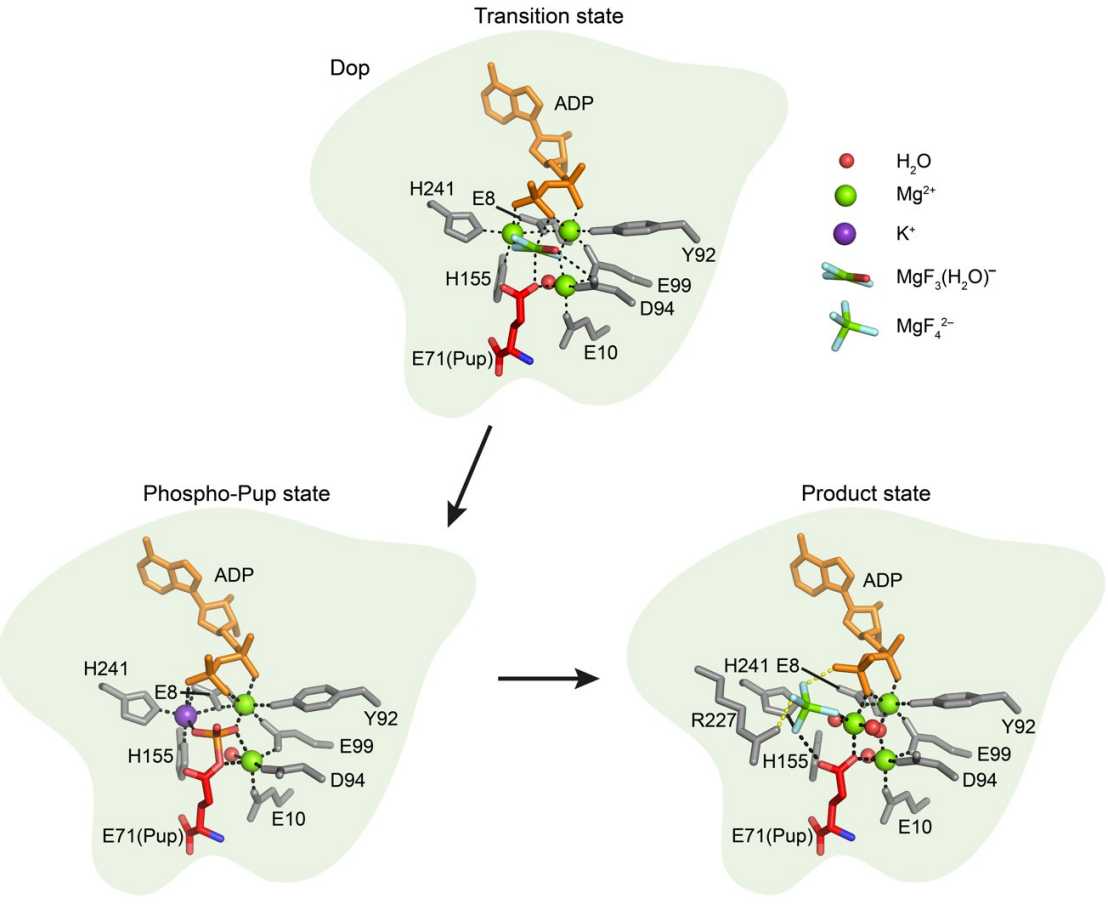
Pupylation is the post-translational modification of lysine side chains on proteins with prokaryotic ubiquitin-like protein (Pup) to target them for proteasomal degradation in mycobacteria and other Actinobacteria. Pupylation supports persistence of Mycobacterium tuberculosis in host macrophages making the enzymes involved in this pathway promising targets for drug development. Pup ligase PafA and depupylase Dop catalyze opposing activities, although they are close structural and sequence homologs and share a common evolutionary origin. In the study published recently in the Journal Nature Communications, Hengjun Cui, with support from colleagues in the Weber-Ban and Ban groups, determined a series of high-resolution crystal structures of Dop in different functional states along the reaction pathway, including Pup-bound states in distinct conformations. Together with biochemical experiments, the structures provide a mechanistic description of the process that generates the catalytic phosphate in the active site of Dop. Formation of the catalytic phosphate from ATP enables Dop to cleave the isopeptide bond between Pup and target proteins and is therefore a distinguishing feature between the ligase and the depupylase. The C-terminal side chain of Pup is directly involved in ATP cleavage and presents the acceptor group in the transition state for phosphoryl transfer. Furthermore, a characteristic loop that is present only in the depupylase enzyme but absent in the ligase supports ATP cleavage. In addition to a mechanistic understanding of depupylation catalyzed by Dop, the study also provides insights into the evolutionary path leading to depupylase versus ligase activities in a pair of enzymes with a common ancestor.
Link to the paper in external page Nature Communications