Ribosomally derived lipopeptides containing distinct fatty acyl moieties
In a recent "PNAS" paper the Piel group (IMB) in collaboration with the Gugger (Institut Pasteur, FR) and Morinaka (National University of Singapore) groups report the first ribosomal peptides with various posttranslational fatty acylations, opening up a synthetic biology route to gene-encoded lipopeptide antibiotics.
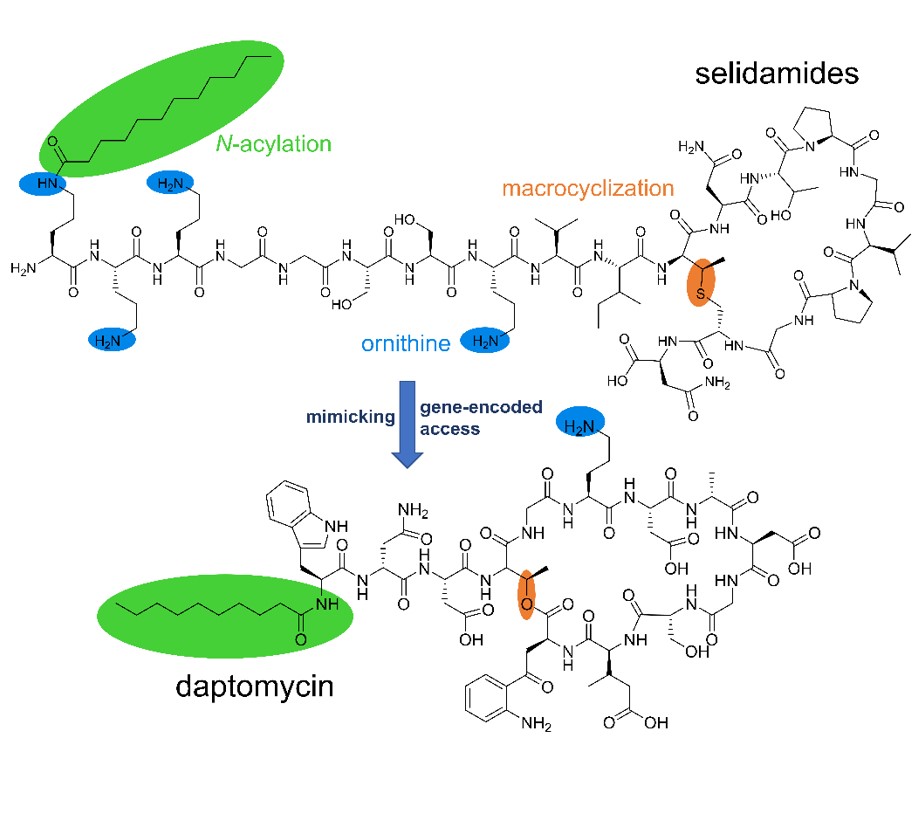
Lipopeptides represent a large group of microbial natural products that include important antibacterial and antifungal drugs and some of the most-powerful known biosurfactants. The vast majority of lipopeptides comprise cyclic peptide backbones N-terminally equipped with various fatty acyl moieties. The known compounds of this type are biosynthesized by nonribosomal peptide synthetases, giant enzyme complexes that assemble their products in a non–gene-encoded manner.
Here, we report the genome-guided discovery of ribosomally derived, fatty-acylated lipopeptides, termed selidamides. Heterologous reconstitution of three pathways, two from cyanobacteria and one from an arctic, ocean-derived alphaproteobacterium, allowed structural characterization of the probable natural products and suggest that selidamides are widespread over various bacterial phyla. The identified representatives feature cyclic peptide moieties and fatty acyl units attached to (hydroxy)ornithine or lysine side chains by maturases of the GCN5-related N-acetyltransferase superfamily. In contrast to nonribosomal lipopeptides that are usually produced as congener mixtures, the three selidamides are selectively fatty acylated with C10, C12, or C16 fatty acids, respectively. These results highlight the ability of ribosomal pathways to emulate products with diverse, nonribosomal-like features and add to the biocatalytic toolbox for peptide drug improvement and targeted discovery.
Link to the paper in external page PNAS