Structure of substrate-translocating ATPase-proteasome complex from M. tuberculosis
A recent publication from the Weber-Ban group (IMBB) published in Nature Communications sheds light on Mycobacterium tuberculosis proteasome substrate engagement and translocation, and reveals a previously unknown interaction element between the proteasomal regulator Mpa and the 20S proteasome gate.
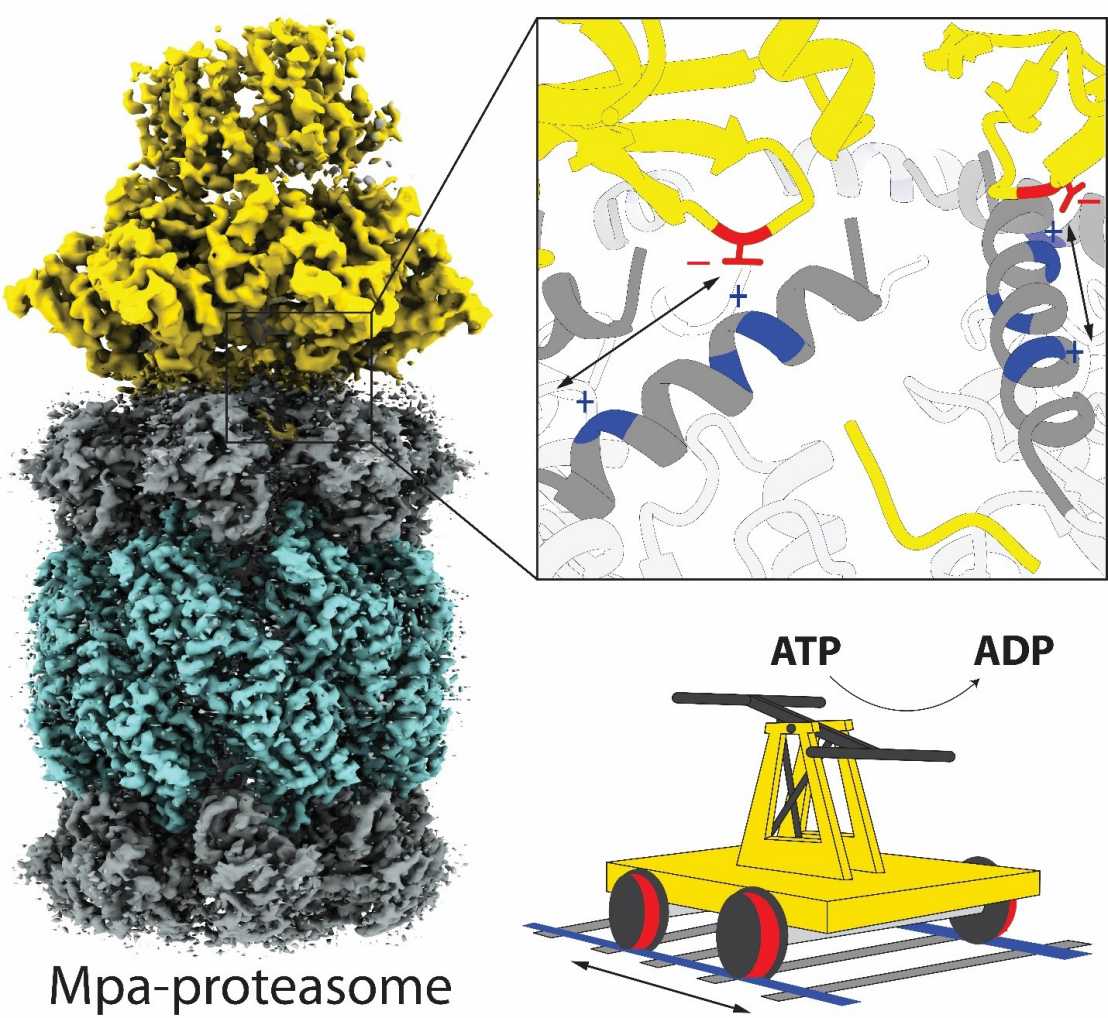
Proteasomes are multi-subunit, molecular machines that carry out controlled protein turnover in the cell. They exist in eukaryotes, archaea and Actinobacteria, including Mycobacterium tuberculosis (Mtb), one of the most successful human pathogens, where proteasomal degradation supports persistence inside the host. Actinobacteria have evolved a ubiquitin-like, post-translational modification pathway where prokaryotic ubiquitin-like protein (Pup) is conjugated to target proteins and serves to direct them for recruitment by the mycobacterial proteasome ATPase (Mpa).
In their study, the authors captured the Mpa-proteasome complex at the crucial stage of engaging and translocating pupylated substrate and visualized its structure in two sequential states of translocation using single-particle cryo-electron microscopy. These results provide a molecular description of how Pup engages with the Mpa pore to initiate substrate processing, and reveals the dynamic character of the interaction between the unfoldase ring and the proteasome core cylinder. The authors identify an interaction element on Mpa that engages with rail-like features present at the proteasomal gate through a series of charge-complementary contacts, allowing Mpa to maintain contact to the 20S despite large conformational changes in the ATPase ring during substrate translocation.
Link to the paper in external page "Nature Communications".