Microbial community assembly is structured by metabolic cross-feeding networks
A recent “Science Advances” paper by the Sauer group (IMSB) uses a model chitin degrading community to demonstrate how metabolic cross-feeding networks are formed by specialized chitin degraders, and how these networks structure the downstream assembly of metabolically diverse populations.
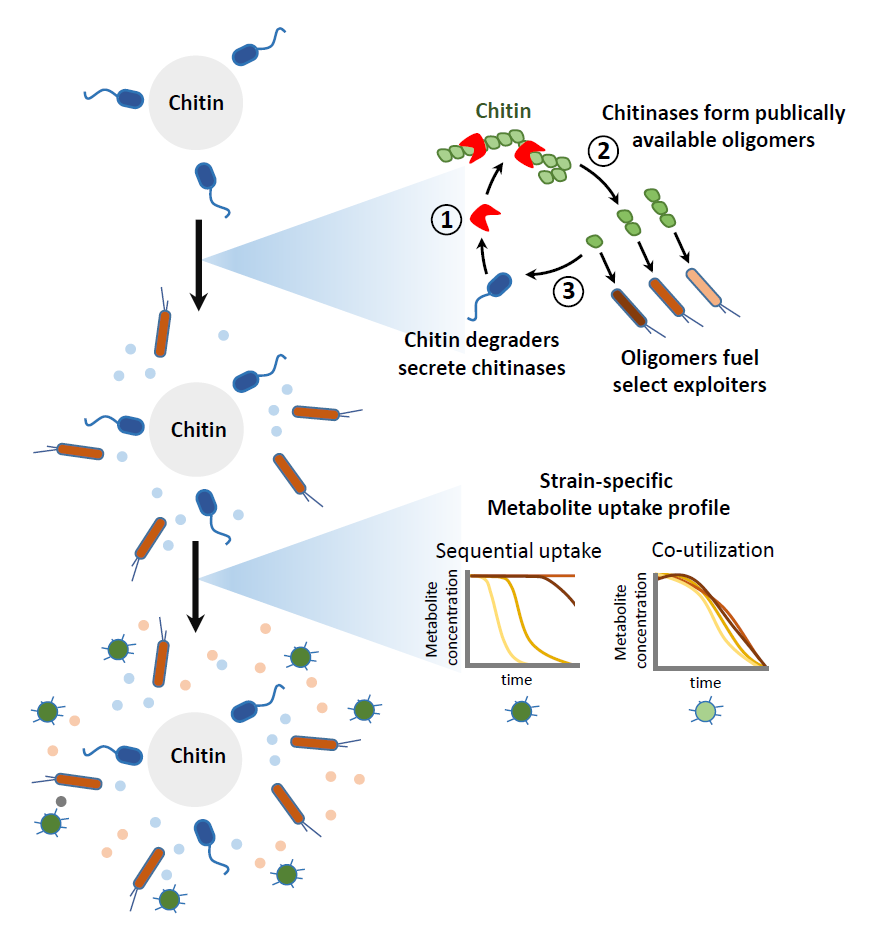
Microbial communities are ubiquitously composed of metabolically diverse populations, even in environments where nutrients are accessible to only small fractions of specialized species. This suggests that metabolic cross-feeding networks form within these communities to provide nutrients to non-specialized species. Yet, it is unclear how these cross-feeding networks form, whether the metabolic activity of specialized species can influence other species that colonize within the community, and how such diverse populations can coexist in the same environment.
We used a model chitin-degrading community to explore these uncertainties and identified different types of metabolic-crossfeeding. Specialized degraders that secrete chitinases first degrade the polysaccharide, but different chitinase activities result in chitin oligomers of different chain lengths. This influences who can colonize, because less specialized consumers are able to consume some of these oligomers and not others. We also demonstrate that all species have distinct preferences for which metabolites they consume, and the order in which they consume these metabolites. Ultimately these preferences determine community composition and dynamics, providing us with some understanding of resources partitioning. Overall, these results demonstrate different ways that metabolites are exchanged in communities to support metabolically diverse populations, yet also demonstrate how metabolic-crossfeeding can determine which species thrive in a community.
Link to the paper in external page Science Advances