Uncovering the interaction landscape of bacteria on Arabidopsis leaves
Published recently in "Nature Microbiology", Schäfer and collaborators (Vorholt lab) systematically probed several thousand interactions among the bacterial leaf microbiota in planta. They found that competition is prevalent and identified a novel peptidase that triggers cell lysis as an interaction mechanism.
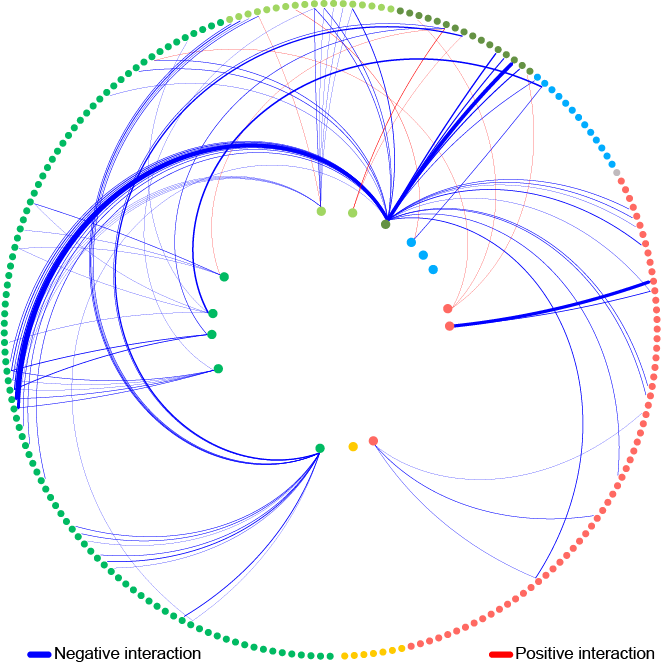
Plants are colonized by a variety of microorganisms dominated by bacteria. Some of these bacteria have beneficial effects on plant health and productivity. However, it remains a challenge to specifically introduce these beneficial bacteria into bacterial communities in a stable manner. Therefore, it is important to understand the factors that determine the success of individual bacteria in invasion and maintenance in the community.
To understand the fundamental drivers of bacterial community assembly, researchers at the Institute of Microbiology systematically probed how a model community of 15 bacteria responds when an additional bacterial strain is added to the community. Quantifying the effects of 200 representative members of the endogenous microbiome, they found that competitive interactions dominate. They then further characterized a pair of strains and discovered that the observed strong reduction in abundance by two orders or magnitude caused by one strain was due to cell lysis that was mediated by a secreted peptidase.
This work unraveled the interaction landscape between bacteria on leaf surfaces and will enable to characterize further mechanisms by which bacteria interact with each other. Increasing mechanistic understanding of these interactions will allow targeted and lasting microbiome engineering to enable crop protection through environmentally friendly approaches.
Link to the paper in external page "Nature Microbiology"