Widespread microbial utilization of ribosomal β-amino acid-containing peptides and proteins
A recent collaborative effort of the Piel (D-BIOL), Bode (D-CHAB) and Zhang (Fudan University) labs was recently published in Chem. It reveals the existence of a widespread family of microbial β-amino acid-containing ribosomal peptides called spliceotides that are highly potent protease-inhibitors.
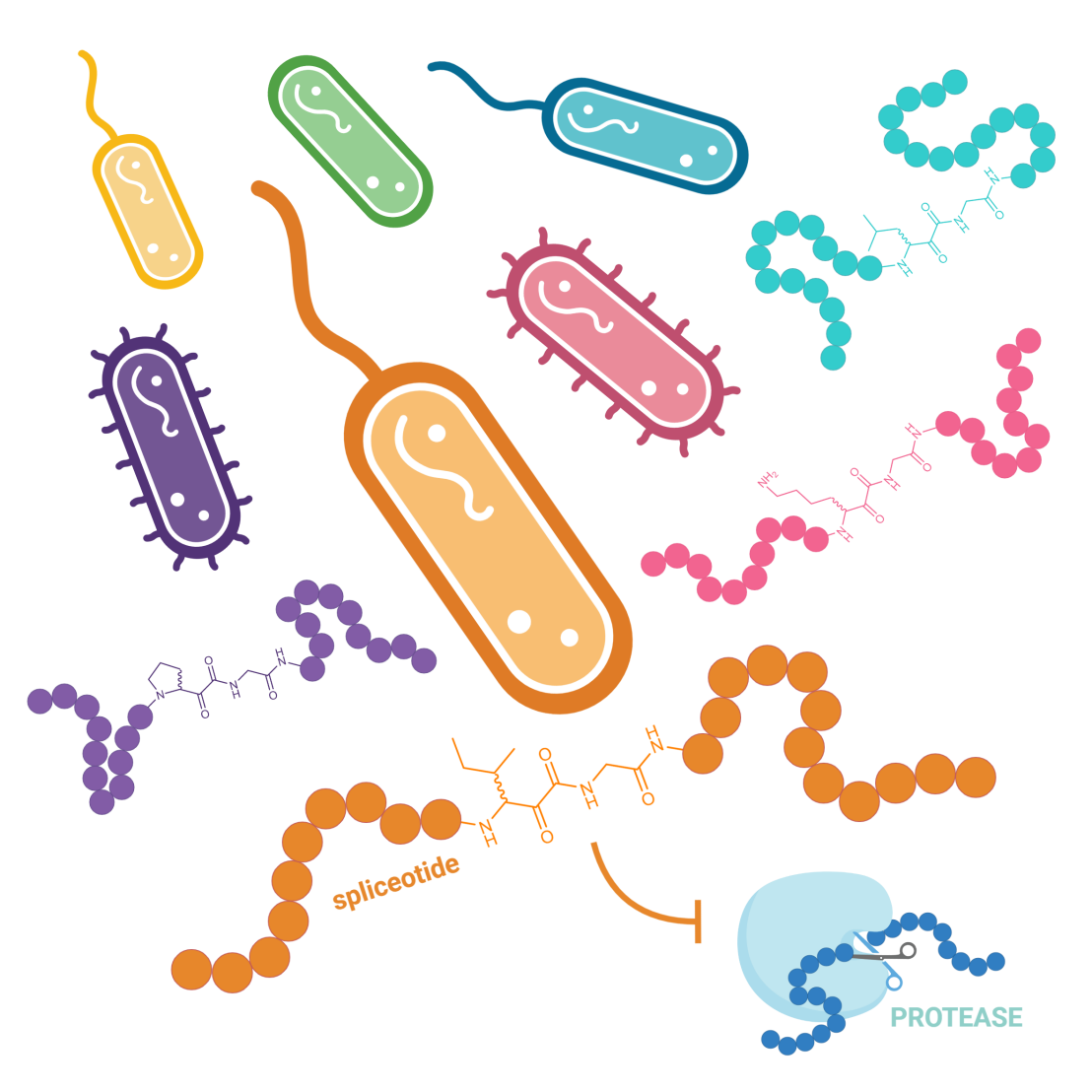
Radical splicease enzymes perform a remarkable post-translational modification that incorporates diverse β-amino acids (ketoamides) into ribosomally synthesized peptides and proteins. Our work reveals its widespread occurrence in bacteria, highlighting a common mechanism to overcome the biosynthetic limitations of the ribosome, which utilizes α-amino acids. We show that the “spliceotides” exhibit potent protease inhibitory activity imparted by the ketoamide moieties, which may benefit the producers in diverse ways. The same β-building blocks are key pharmacophores in synthetic protease inhibitors, including the SARS-CoV-2 protease. Nature, then, has already evolved an enzymatic route to high-value pharmacophores that can be challenging to access synthetically. This provides a biosynthetic, gene-encoded toolbox that offers access to at least 15 different β-amino residues, substantially expanding the scope of ribosomal products and representing an invaluable resource for drug discovery.
Link to the paper in external page Chem.