Towards a structurally resolved human protein interaction network
A recent "Nature Structural and Molecular Biology" paper by the Beltrao group (IMSB) in collaboration with the Elofsson group (Stockholm University, Sweden) have predicted complex structures for 65,000 pairs of human proteins and identify interfaces harbouring disease mutations.
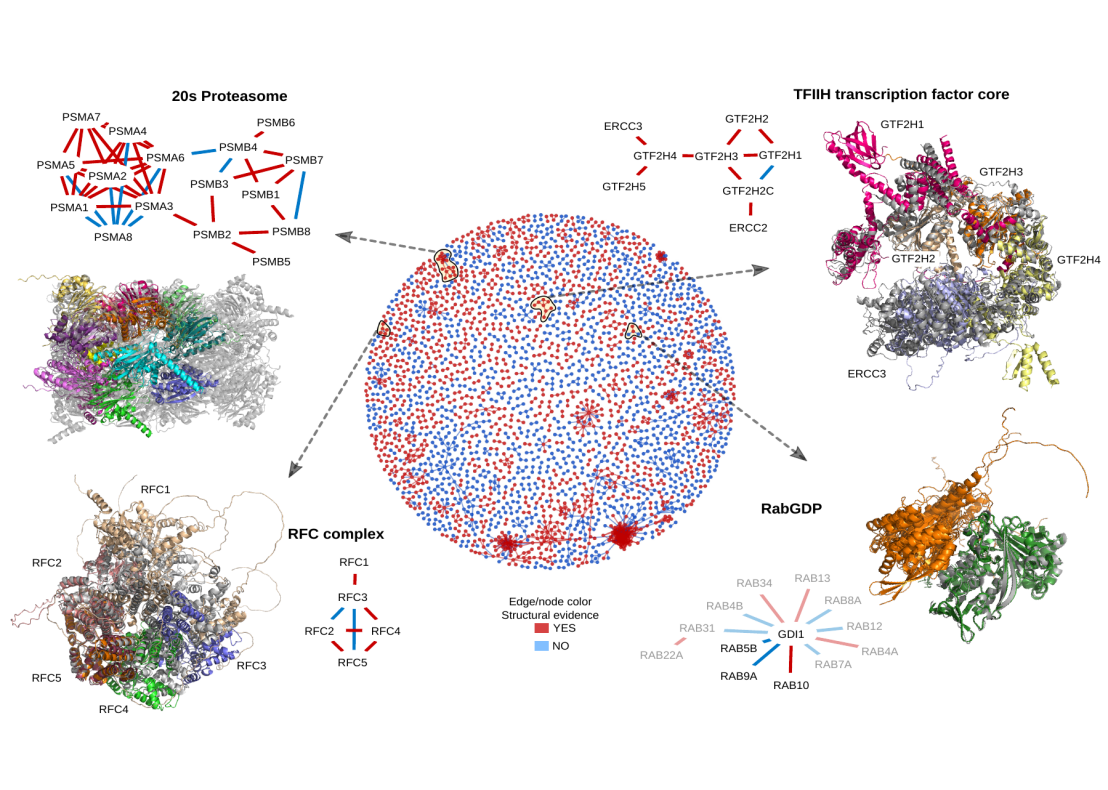
Proteins are key cellular effectors determining most cellular processes. These rarely act in isolation, but instead, the coordination of the diversity of processes arises from the interactions among multiple proteins and other biomolecules. The characterization of protein-protein interactions is crucial for understanding which groups of proteins form functional units and underlies the study of the biology of the cell. The structural characterisation of protein complexes is a critical step in understanding the mechanisms of protein function, studying the impact of mutations and the regulation of cellular processes.
Researchers at IMSB have applied recent advances in machine learning to predict complex structures for 65,000 pairs of human proteins known to physically interact. They identify over three thousand high-confidence models, of which nearly half have no homology to a known structure. They also identify interface residues harbouring disease mutations, suggesting potential mechanisms for pathogenic variants. These predicted binary complexes can be used to build larger assemblies helping to expand our understanding of human cell biology and disease processes.
Link to the paper in external page "Nature Structural and Molecular Biology".