Electrostatic interactions guide substrate recognition of the prokaryotic ubiquitin-like protein ligase PafA
A recent "Nature Communications" paper by the Weber-Ban group (IMBB) investigated how the single ligase PafA of the pupylation pathway interacts with its protein substrates, demonstrating how PafA can achieve recognition of a wide array of substrates while retaining selective protein turnover.
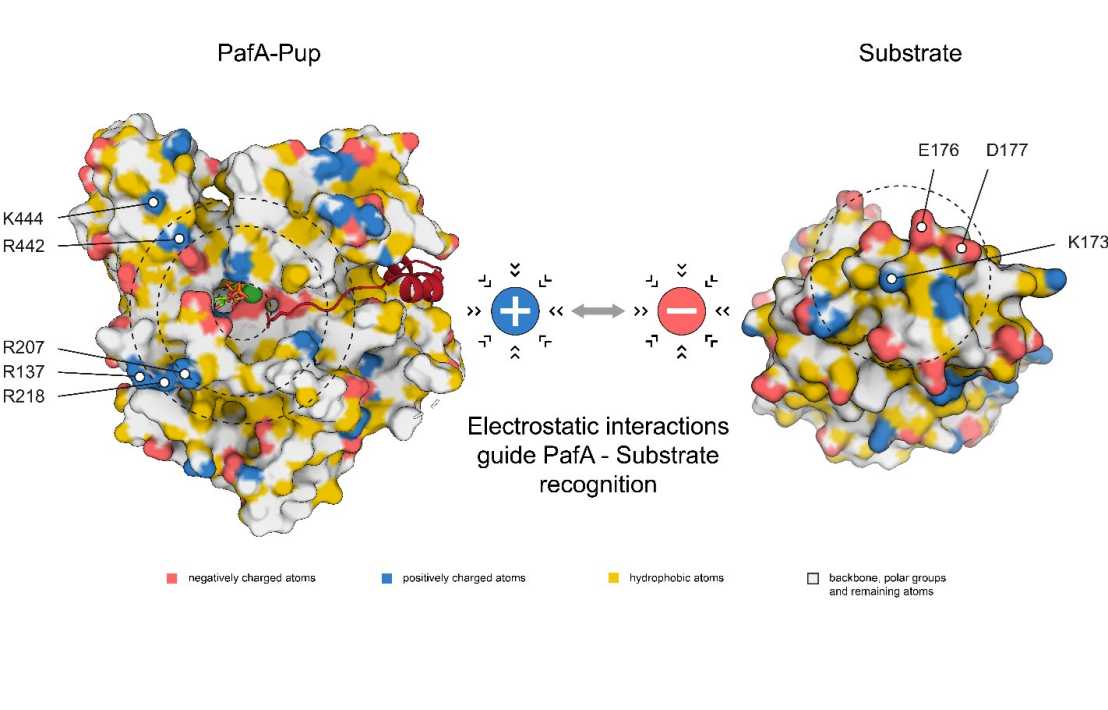
Regulated protein degradation by chaperone-proteases is a fundamental process in all living organisms. In addition to the usual bacterial degradation complexes (e.g. Clp proteases), Actinobacteria possess a proteasome assembly related to eukaryotic proteasomes. However, instead of ubiquitination that marks substrates for degradation by the eukaryotic proteasome, Actinobacteria have developed their own targeting system in the form of pupylation. Here, the small protein Pup is attached to substrates by the single ligase PafA to mark them for degradation. Nevertheless, hundreds of potential substrates have been identified raising the question of how a single enzyme is capable of recognizing a range of substrates while retaining substrate specificity.
Researchers in the Weber-Ban group identified several highly conserved basic residues at the rim of the active site of PafA that are required to effectively pupylate different substrates. Additionally, they located a mirroring tertiary structure feature in the form of negatively charged residues distributed around the target pupylation site. By probing potential electrostatic interactions between ligase and substrate, they were able to elucidate the selection mechanism of PafA that allows for substrate specificity and subsequently ensuring selective protein turnover.
Link to the paper in external page "Nature communications".