Animal-associated marine Acidobacteria with a rich natural product repertoire
A recent ”Chem” paper by the Piel and Sunagawa groups (IMB) in collaboration with the Wakimoto lab at Hokkaido University reveals Acidobacteria, a poorly studied bacterial phylum, as a rich source of bioactive natural products.
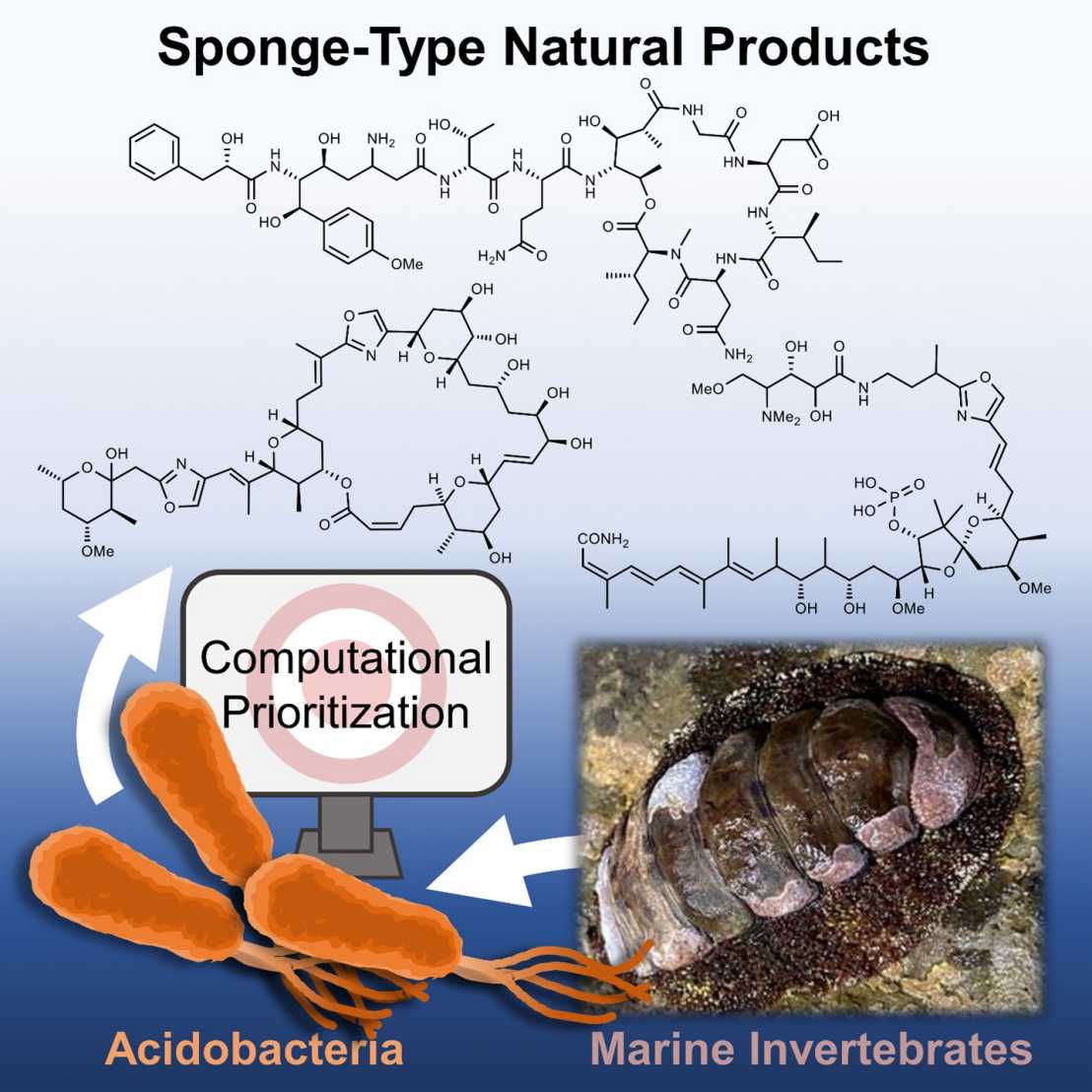
Marine sponges contain numerous natural products with diverse and important pharmacological activities. However, the limited supply and non-sustainable animal harvesting are major hurdles for drug development. Previous studies have shown that many natural products are produced by uncultivated bacterial symbionts and that their biosynthetic genes are commonly transferred between distantly related bacteria.
In this study, we applied these insights to a computational strategy to identify alternative, culturable producers by compound prediction from bacterial genomes. This strategy predicted the acidobacterial family Acanthopleuribacteraceae as an untapped resource enriched in sponge-type and new chemistry, which was validated by the isolation of several bioactive polyketides and peptides.
Additional sequence analyses of marine animal microbiomes indicated that cultivation efforts targeting Acanthopleuribacteraceae could be rewarded with a multitude of further valuable natural products.
In brief:
- Computational pipeline identifies rich and sustainable sources of valuable chemistry
- Acanthopleuribacteraceae produce sponge-type natural products
- Isolation and structure elucidation of ten natural products from three pathways
- First natural products from Acidobacteria, a ubiquitous bacterial phylum
Link to the paper in external page "Chem".