Conformations of Macrocyclic Peptides Sampled by NMR: Models for Cell-Permeability
A recent “Journal of the American Chemical Society” paper by the biomolecular NMR platform (BNSP) in collaboration with the Künzler group (Institute of Microbiology) and Prof. Güntert (IMPS, D-CHAB) presents a novel approach to probe conformations and dynamics of peptides.
The biological activities and pharmacological properties of peptides and peptide mimetics are determined by their conformational states. Therefore, a detailed understanding of the conformational landscape is crucial for rational drug design. Nuclear magnetic resonance (NMR) is the only method for structure determination in solution.
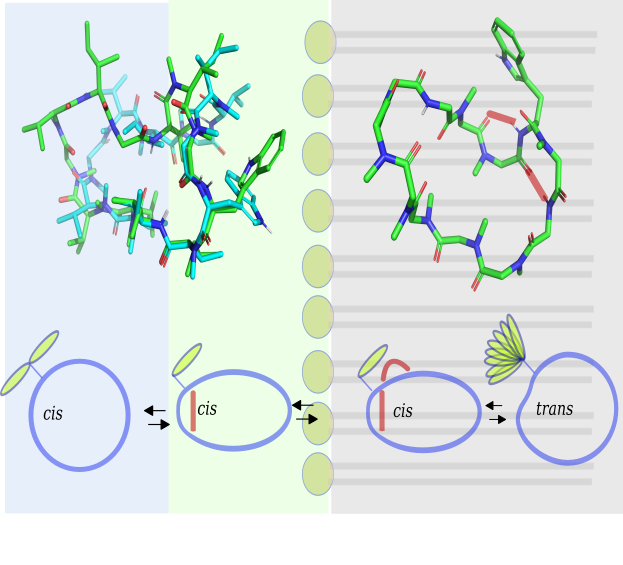
In this study, we introduce a new approach to investigating the structures of modified macrocyclic peptides. Exact NOEs (eNOEs) in viscous solvent mixtures are used to replicate various cellular environments. eNOEs provide detailed structural information for highly dynamic modified peptides. Structures of high precision were obtained for cyclosporin A, with a backbone atom rmsd of 0.10 Å. Distinct conformational states in different environments were identified for omphalotin A (OmphA), a fungal nematotoxic and multiple backbone N-methylated macrocyclic peptides.
A model for cell-permeation is presented for OmphA, based on its structures in polar, apolar, and mixed polarity solvents. During the transition from a polar to an apolar environment, OmphA undergoes a rearrangement of its H-bonding network, accompanied by a cis to trans isomerization of the ω torsion angle within a type VIa β-turn. We hypothesize that the kinetics of these conformational transitions play a crucial role in determining the membrane-permeation capabilities of OmphA.
Link to the paper in the external page "Journal of the American Chemical Society".