Phylogenetic inference reveals clonal heterogeneity in circulating tumor cell clusters
In a recent "Nature genetics" paper, the Aceto group (IMHS) in collaboration with the Beerenwinkel group (D-BSSE) interrogate clonal heterogeneity in individual circulating tumor cell clusters (CTC clusters), thereby improving our understanding of metastasis-relevant clonal dynamics.
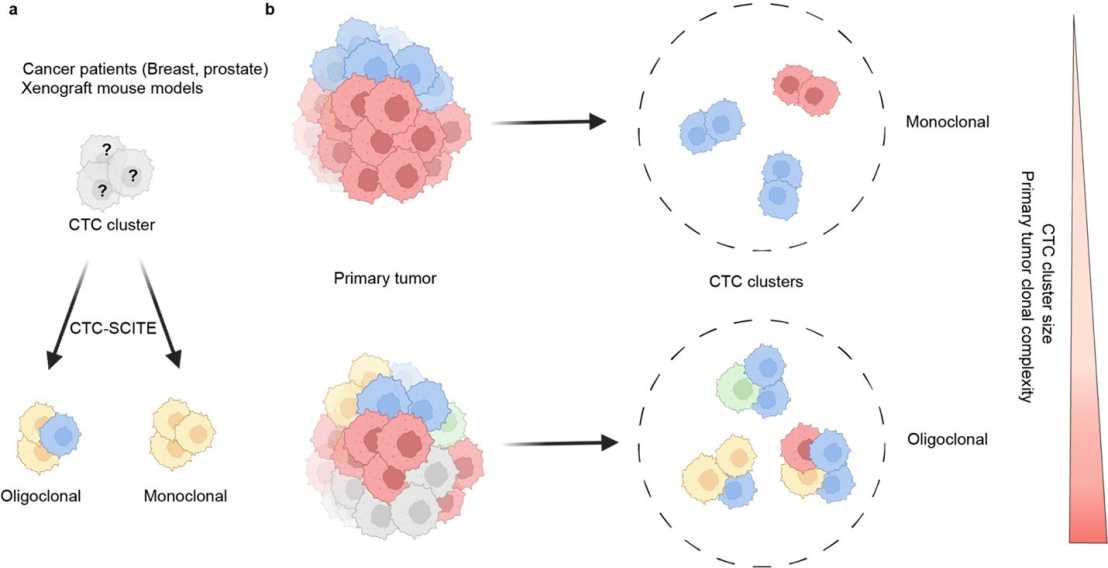
Circulating tumor cell clusters (CTC clusters) are highly efficient metastatic seeds in various cancers. Yet, their genetic heterogeneity and clonal architecture is poorly characterized.
Researchers at the IMHS and D-BSSE developed a pioneering method (CTC-SCITE) to infer the clonality of individual CTC clusters isolated from patients with breast and prostate cancer, as well as breast cancer xenograft mouse models. The study provides direct evidence for the existence of individual CTC clusters carrying cells from different tumor clones, proposing them as key contributors of genetic diversity in metastasis and as promising targets when aiming to suppress the spread of genetically divergent secondary tumor lesions. Using barcode-mediated clonal tracking in vivo, the authors link the frequency of heterogeneous CTC clusters to primary tumor clonal diversity and cluster size. These findings suggest that quantitative assessment of the clonal composition of CTCs in liquid biopsies could provide insights about the genetic diversity of corresponding primary or metastatic lesions, potentially mitigating the spatial and temporal limitations of traditional tissue biopsies.
Link to the paper in external page "Nature Genetics"