Phosphorylation-linked complex profiling identifies assemblies required for Hippo signal integration
A recent Molecular Systems Biology paper by the Gstaiger & Aebersold group (IMSB) in collaboration with the Peter group (IBC) developed a new proteomic workflow to study the functional relationships between PTMs and complex formation.
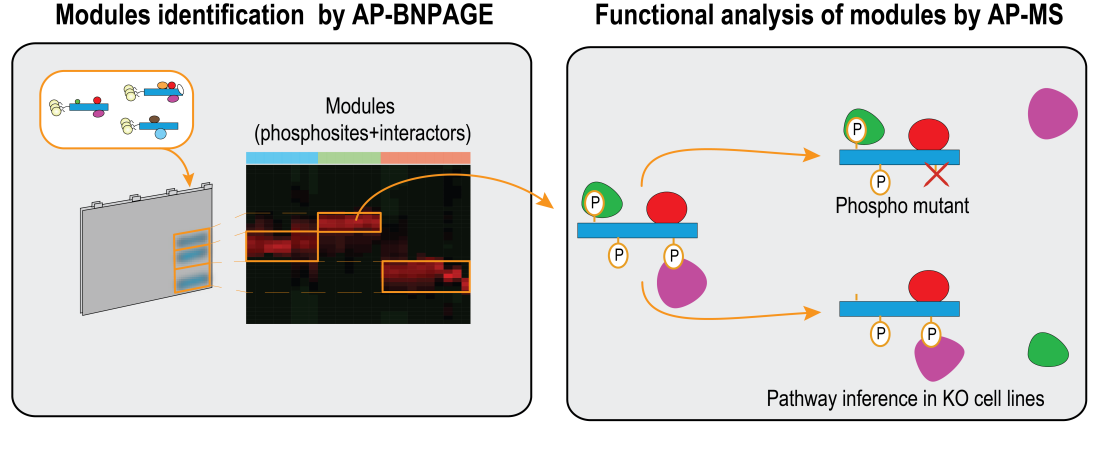
While several computational methods have been developed to predict the functional relevance of phosphorylation sites, experimental analysis of the interdependency between protein phosphorylation and Protein–Protein Interactions (PPIs) remains challenging. Here, we describe an experimental strategy to establish interdependencies between protein phosphorylation and complex formation. This strategy is based on three main steps: (i) systematically charting the phosphorylation landscape of a target protein; (ii) assigning distinct proteoforms of the target protein to different protein complexes by native complex separation (AP-BNPAGE) and protein correlation profiling; and (iii) analyzing proteoforms and complexes in cells lacking regulators of the target protein.
We applied this strategy to YAP1, a transcriptional co-activator for the control of organ size and tissue homeostasis that is highly phosphorylated and among the most connected proteins in human cells. We identified multiple YAP1 phosphosites associated with distinct complexes and inferred how both are controlled by Hippo pathway members. We detected a PTPN14/LATS1/YAP1 complex and suggest a model how PTPN14 inhibits YAP1 via augmenting WW domain-dependent complex formation and phosphorylation by LATS1/2.
Link to the paper in external page "Molecular Systems Biology".