ClpC2 protects an essential mycobacterial degradation pathway against cyclomarin A toxicity
In a recent paper published in “Communications Biology”, the Weber-Ban group (IMBB) demonstrates a role for a ClpC1 partial homologue in helping to protect an essential protein degradation pathway against the antibiotic Cyclomarin A in Mycobacterium tuberculosis.
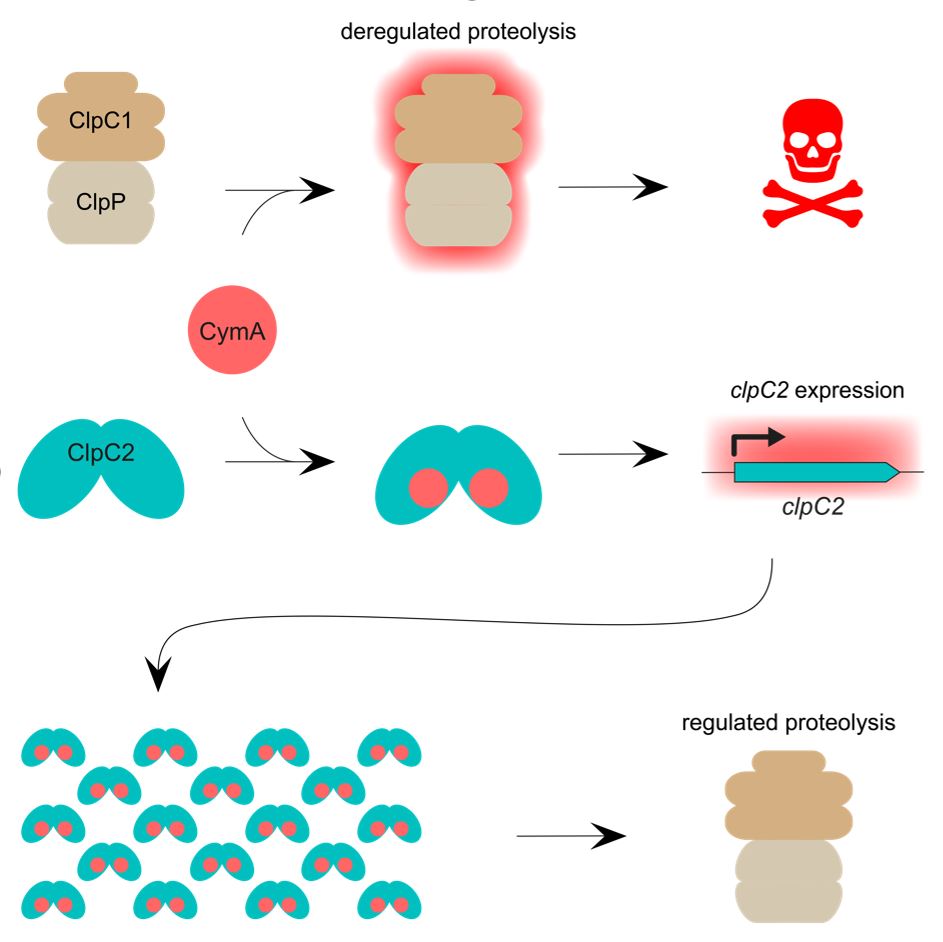
In the 20th century, the advent of antibiotics was revolutionary in the treatment of tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), but this has since been undermined by the rise of multi-drug resistant TB. We therefore need new antibiotics with novel modes of action. The Mtb Clp chaperone-protease complexes have garnered interest as potentially promising drug targets. Interestingly, several natural products with antitubercular activity have been found to target the chaperone ClpC1 including Cyclomarin A (CymA).
Researchers at IMBB reveal the ClpC1 partial homologue, ClpC2, also binds to the antibiotic CymA in Mtb. Moreover, structural studies were combined with biochemical and microbiological approaches to demonstrate that ClpC2 helps to protect mycobacteria against CymA-induced toxicity by competing with ClpC1 for CymA. Further investigations show ClpC2 cellular concentration increases considerably upon exposure to CymA and this upregulation is directed at a transcriptional level. Additionally, ClpC2 is found to have a role in regulating its own transcription in response to CymA. Thus, ClpC2 acts as a macromolecular sponge to prevent CymA interference in ClpC1-mediated protein degradation. The work presented here highlights the importance of ClpC2 to our growing understanding of the Clp complexes as drug targets.
Link to the paper in external page "Communications Biology".