ATP-independent substrate recruitment to proteasomal degradation in mycobacteria
A recent "Life Science Alliance " paper by the Weber-Ban group (IMBB) in collaboration with the Picotti group (IMSB) demonstrates the key determinants for substrate selection for ATP-independent proteasomal degradation, which include but are not limited to partial disorder.
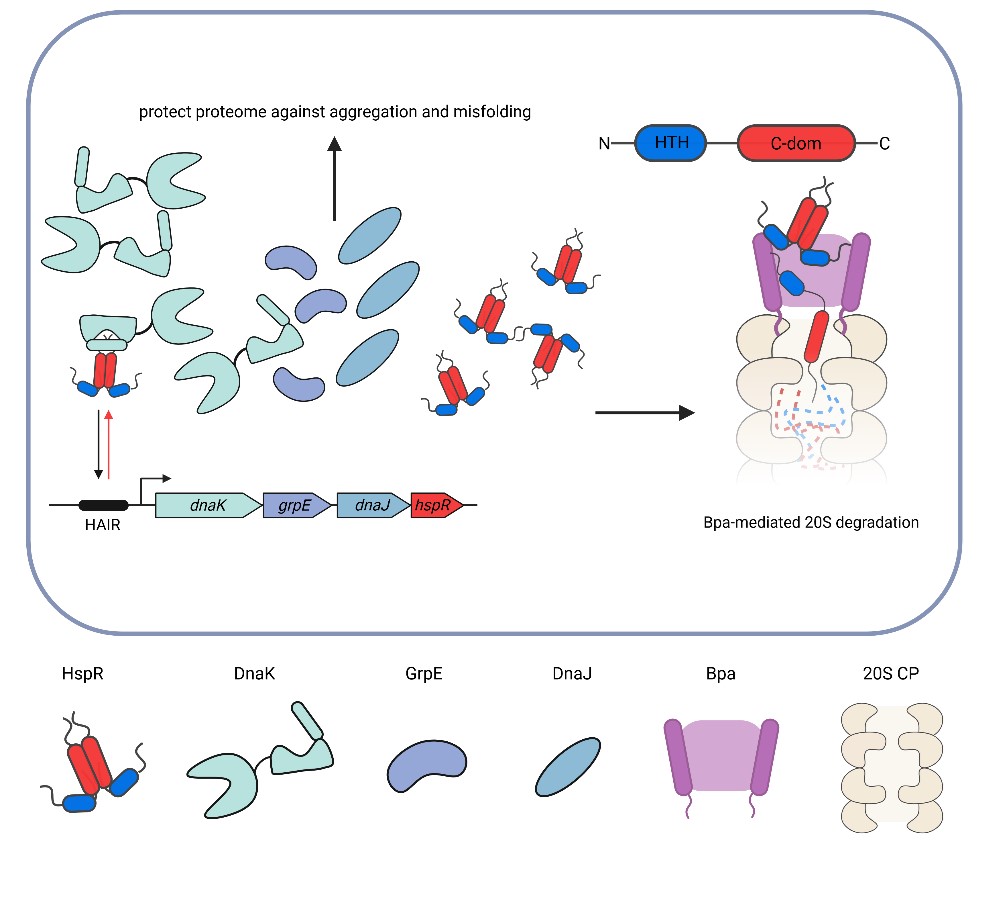
Protein turnover, achieved through controlled protein degradation by compartmentalizing proteases, plays a crucial role in bacteria, serving as a mechanism for quality control and regulation. Mycobacteria have, in addition to various bacteria-typic chaperone-protease systems, acquired a eukaryotic-like proteasome through horizontal gene transfer to support survival under stress, for example during the persistence of the human pathogen Mycobacterium tuberculosis inside host macrophages.
Researchers in the Weber-Ban group have previously identified bacterial proteasome activator Bpa, which facilitates ATP-independent proteasomal degradation. A known natural substrate is the heat shock repressor HspR from M. tuberculosis, which regulates the expression of the Hsp70/40 chaperone system. As Bpa lacks an ATPase motor domain that unfolds substrates for translocation into the proteasomal degradation chamber, the underlying mechanism has remained elusive.
The study suggests a mechanism for Bpa-mediated recruitment of HspR that is not based solely on conformational selection but also involves recognition of folded regions of the substrate. In the first step Bpa binds a region in the N-terminal helix-turn-helix domain of HspR. However, in order to process the substrate, a disordered C-terminal region of HspR is required. Processive translocation of HspR into the degradation chamber likely relies on the dynamic folding/unfolding equilibrium of HspR and further binding events inside the proteasomal degradation chamber.
Link to the paper in external page "Life Science Alliance".