Genomic footprinting uncovers global transcription factor responses to amino acids in E. coli
A "Cell Systems" paper by the Sauer lab (IMSB) used a recent bacterial genomic footprinting approach to globally map the transcriptional response of E. coli to four amino acids, revealing much broader regulatory responses than anticipated, with about 682 novel transcription factor-DNA interactions.
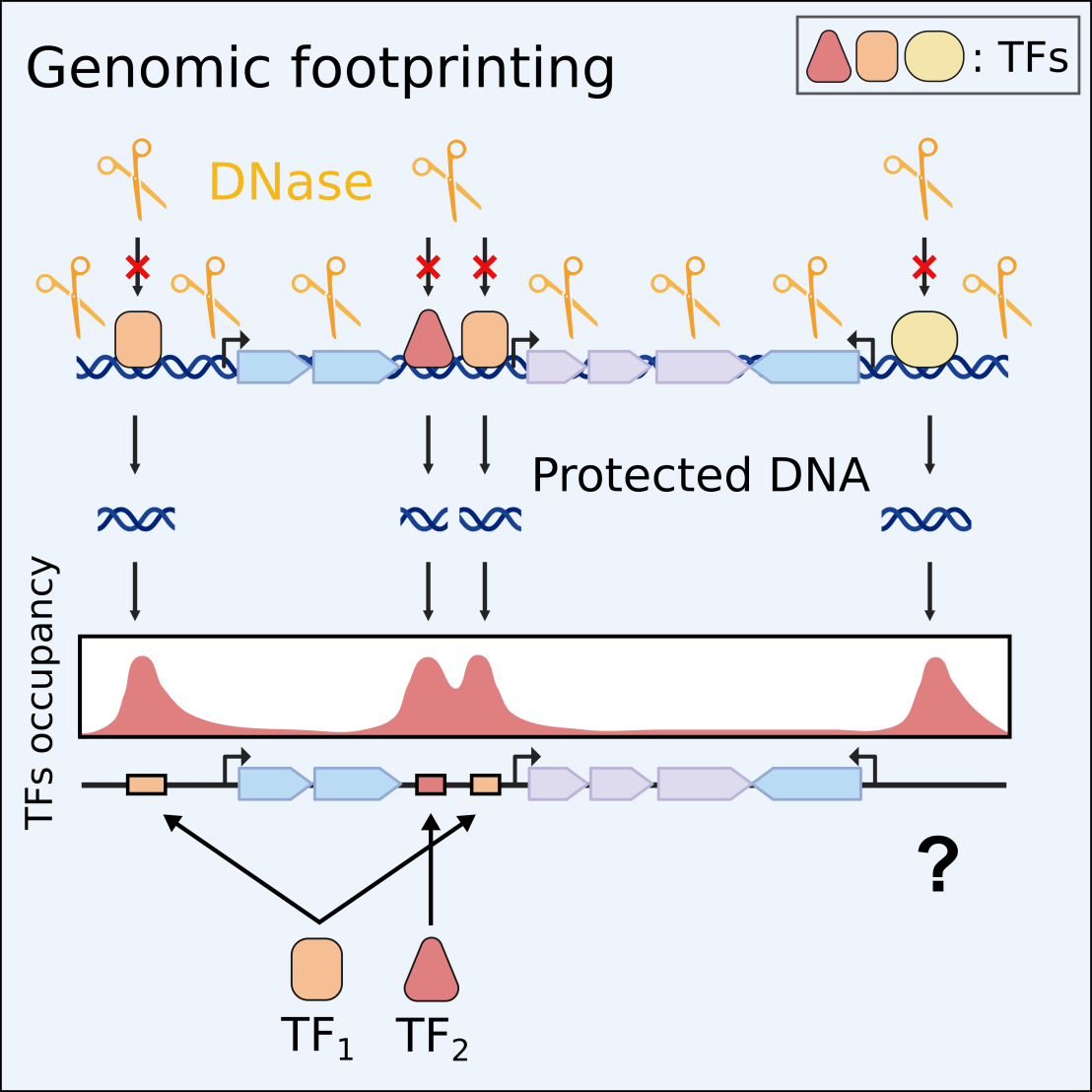
Our knowledge of bacterial regulatory responses comes mainly from few well-studied transcription factors (TFs), often lacking an unbiased genome-wide perspective. Using a recent genomic footprinting approach, we comprehensively mapped the genome-wide regulatory responses of E. coli to leucine, methionine, alanine, and lysine. The global TF Lrp was found to individually sense three amino acids and mount three different target gene responses. Overall, 531 genes had altered RNA polymerase occupancy and 32 TFs responded directly or indirectly to the presence of amino acids, including regulators of membrane and osmotic pressure homeostasis. About 70% of the detected TF-DNA interactions were previously unknown. We thus identified 682 novel TF binding locations, for a subset of which the involved TFs were identified by affinity purification. This comprehensive map of amino acid regulation illustrates the incompleteness of the known transcriptional regulation network, even in E. coli - the best characterized bacterium.
Link to the paper in external page "Cell Systems".