Determination of an SR Protein's RNA Preference Points to its Chaperone Role for Splicing
In a "Nature Communications" paper, the Allain lab (IBC) with the Panse and Robinson groups (UZH) elucidated with a new method the in vivo RNA targets of NPL3, solved the structure of its RRMs with RNA, and then based on this knowledge could show that the protein likely acts an RNA chaperone for spliceosome assembly.
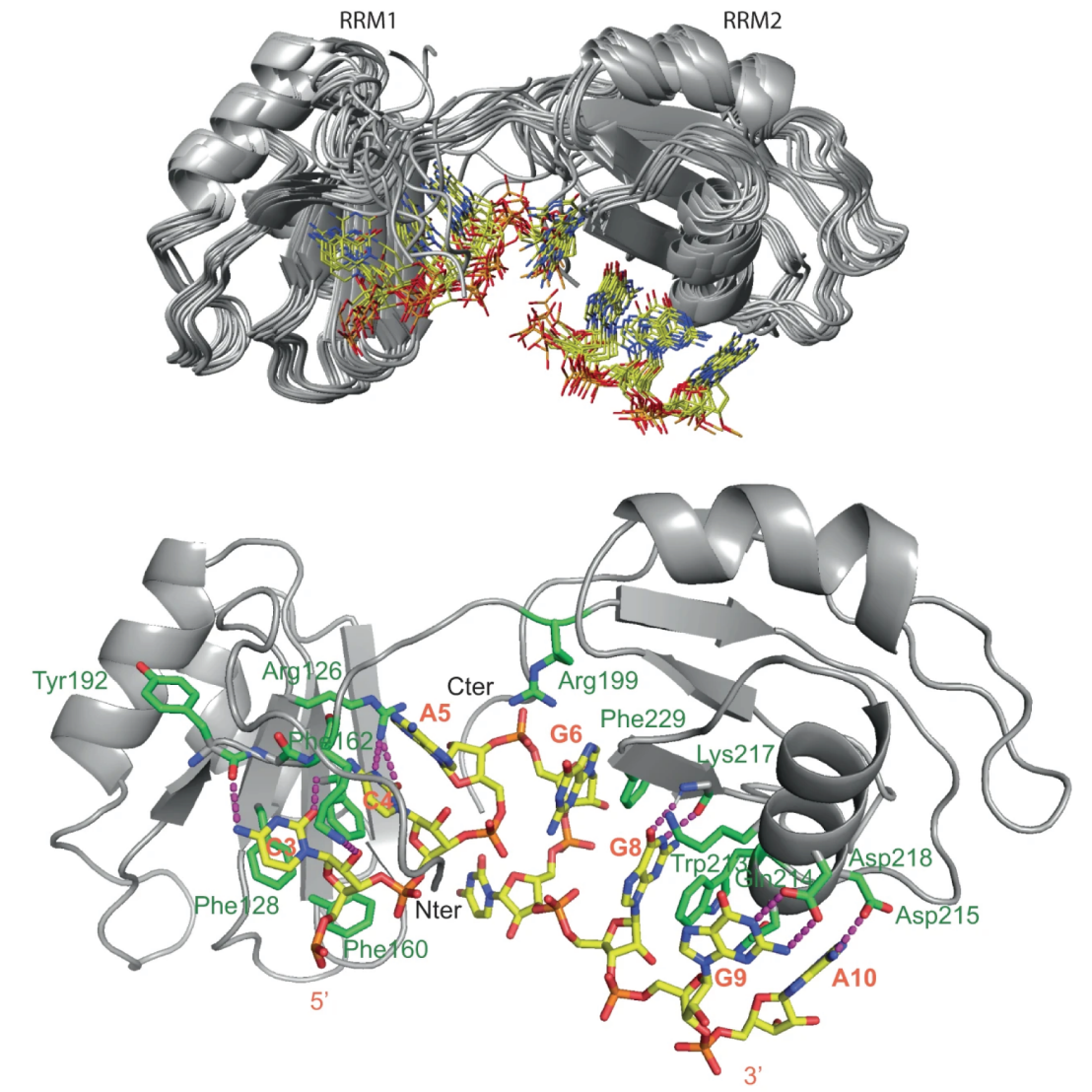
Less than 5% of S. cerevisiae genes undergo splicing and the organism possesses only one Serine and Arginine-rich protein (SR protein) called NPL3 with a known involvement in splicing, including splicing promotion and spliceosome assembly. The protein's in vivo RNA binding preference was unknown despite previous studies. Applying a novel method called split-iCRAC, they could elucidate the in vivo RNA targets and derive a binding consensus sequence.
Based on this consensus sequence, the researchers could determine the complex structure of the two RRMs with RNA, revealing a cooperative mode of RNA binding by the RRMs, also involving the linker between them, leading to a fixed orientation of the two domains. They then used the structural knowledge to derive several binding mutants subsequently assessed biochemically and in in vivo complementation experiments.
These experiments confirmed the protein's involvement in spliceosome assembly and the split-iCRAC data showed a strong enrichment of crosslinks at the 5' end of U2 snRNA, which forms a stem that must be melted in spliceosome assembly to allow duplex formation of U2 and U6 snRNAs. This formation is essential for the formation of the spliceosome's active site, and further experiments provided evidence that NPL3 promotes the melting of this stem, pointing to an unexpected RNA chaperone role of this protein during spliceosome assembly.
Link to the paper in external page "Nature Communications".