Dimerization of a 5 kDa domain defines the architecture of a 5 MDa mega-enzyme complex
In a recent paper in Science Advances, the Glockshuber group (IMBB) solves a 60-year-old conundrum and shows how dimerization of a 5 kDa domain determines the architecture and stoichiometry of the 5 MDa pyruvate dehydrogenase multienzyme complex from Gammaproteobacteria.
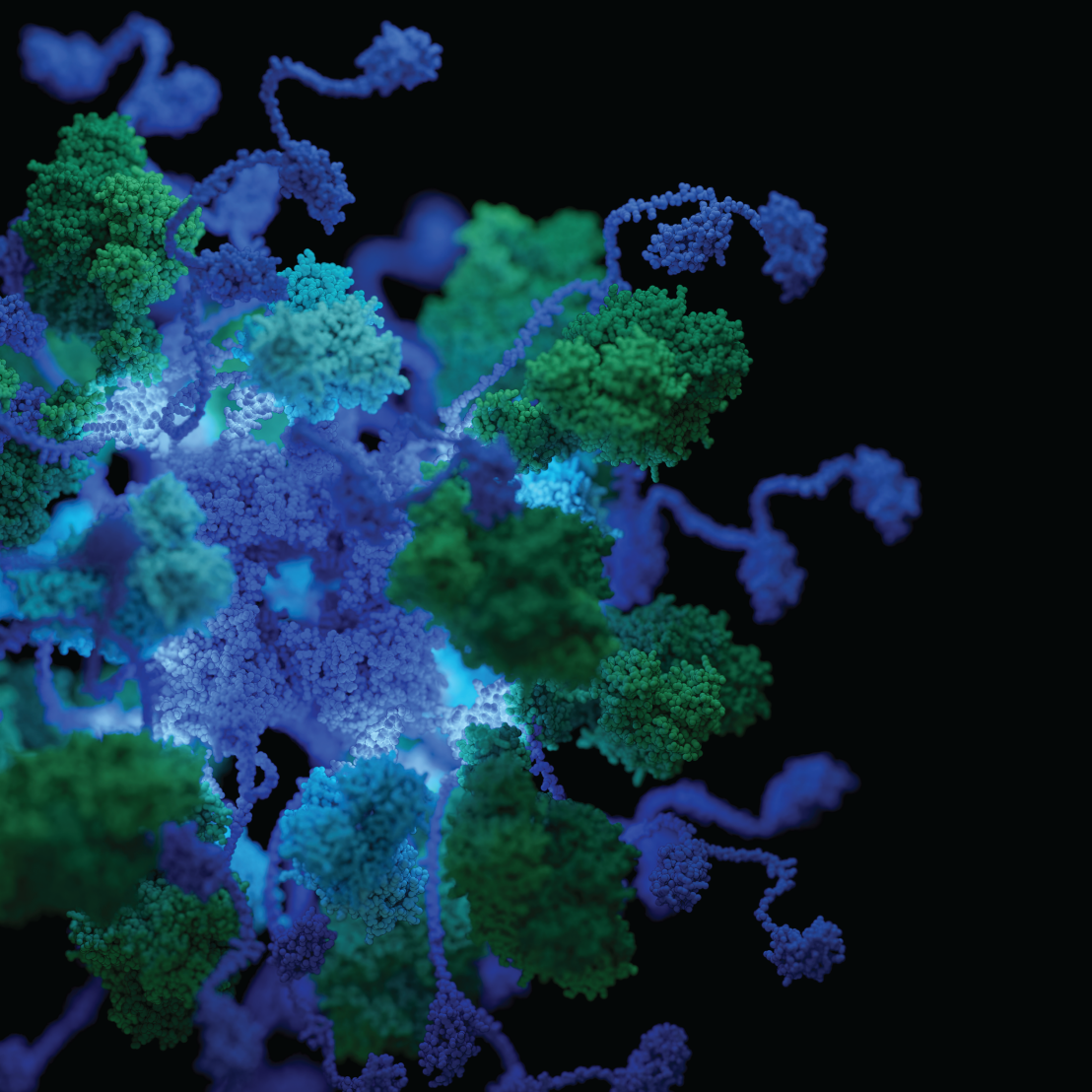
The pyruvate dehydrogenase complex (PDHc) is a key metabolic mega-enzyme, linking glycolysis with the citric acid cycle by converting pyruvate into acetyl-CoA. Its critical role in energy production makes PDHc an integral subject in every biochemistry curriculum. Across all domains of life, PDHcs form large assemblies composed of the subunit dihydrolipoamide acetyltransferase (E2) that forms an oligomeric core complex, and the homodimeric, peripheral subunits pyruvate dehydrogenase (E1) and dihydrolipoamide dehydrogenase (E3) that associate with the core via the peripheral-subunit binding domain (PSBD) of E2.
Using biochemical experiments, X-ray crystallography and electron microscopy, the authors show that the large multienzyme PDHc from E. coli can be simplified to a smaller functional unit termed mini-PDHc. The studies on the mini- and wild-type PDH complexes demonstrate that two of the three PSBDs in homotrimeric E2 dimerize and associate with two E1 dimers. The third, unpaired PSBD in the E2 homotrimer binds exclusively to E3 dimers. The results show that dimerization of the 5 kDa PSBD is the key determinant of the PDHc architecture, guaranteeing accommodation of both peripheral subunits and thus maximum enzymatic activity.
In contrast to the stoichiometry of PDHc described in textbooks (E1:E2:E3 = 24:24:12), the correct stoichiometry of the megacomplex is E1:E2:E3 = 32:24:16, with a total mass of 5.6 MDa. Sequence analyses revealed that this architecture likely applies to PDHcs from all Gammaproteobacteria. This deepens our understanding of this key metabolic mega-enzyme in a wide range of species, including medically relevant human pathogens.
Link to the paper in external page "Science Advances".