Assessing microbiome population dynamics using genomic WISH-tags
To study the dynamics of microbiome assembly with strain-level resolution, the Vorholt, Hardt, and Sunagawa labs (all IMB) jointly developed a novel genomic barcoding system, the WISH-tags. These were used to assess how the order of arrival order impacts colonization in plant and mammalian microbiomes.
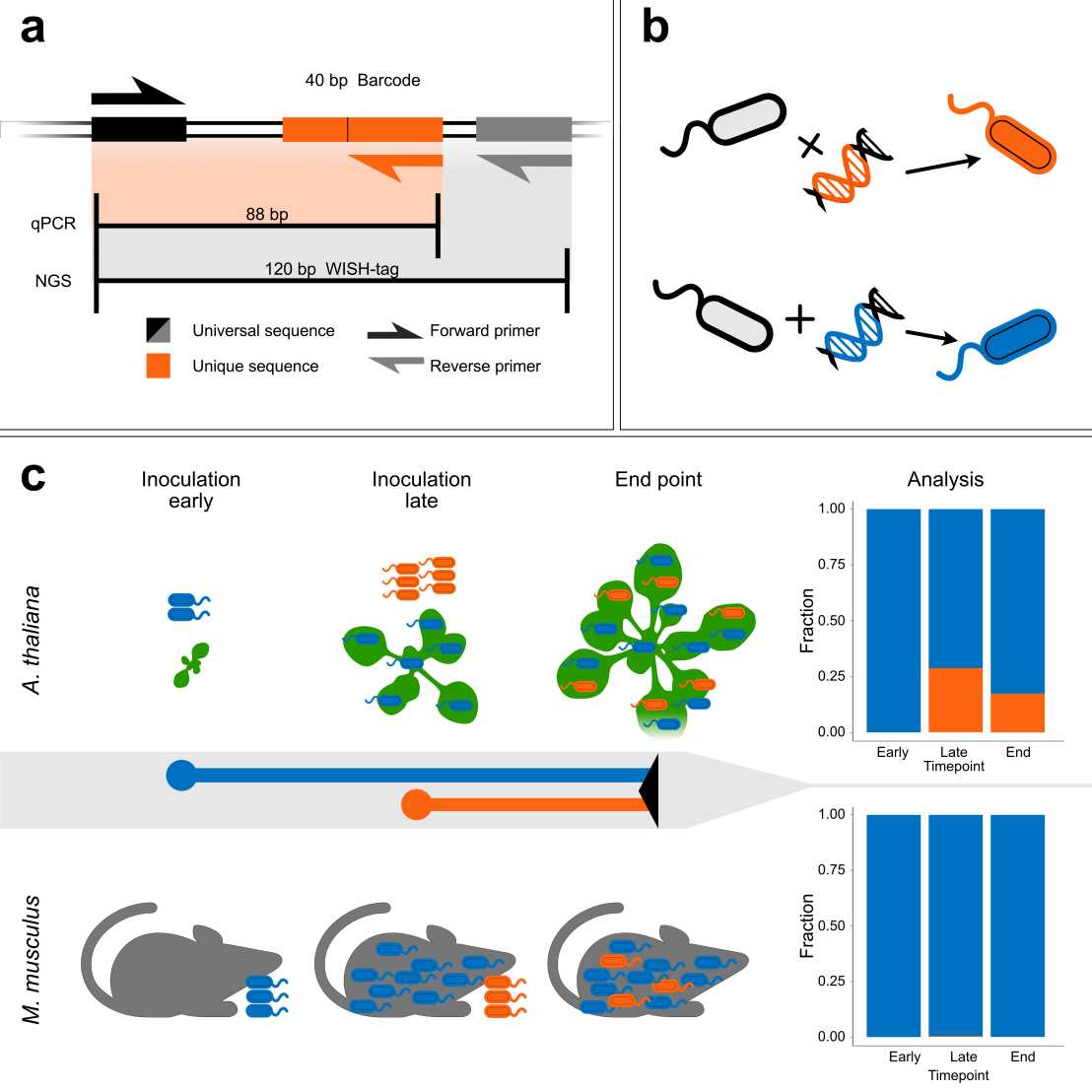
Microbes recurrently assemble into structured microbiomes that play critical roles in plant and animal hosts. However, the observed composition of microbiomes can mask diverse underlying population dynamics, requiring resolution at the intra-strain level. In a recent collaborative study of the NCCR Microbiomes, researchers from the Vorholt, Hardt, and Sunagawa labs developed a novel genomic barcoding system called wild-type isogenic standardized hybrid (WISH)-tags. These tags enable quantification and tracing of microbial populations using qPCR or next-generation sequencing. Daniel et al. introduced the WISH-tags into model and non-model bacteria from the mouse and plant microbiota.
Focusing on two bacterial strains, they investigated intra-strain priority effects in the murine gut and the Arabidopsis phyllosphere. Curiously, the outcome was different in the two hosts. In the gut, late-arriving strains were prevented from establishing new populations, whereas in the phyllosphere, latecomers could still thrive even with an established population present. With their proof-of-concept application the authors showed that WISH-tags are a valuable resource for deciphering the population dynamics underlying microbiome assembly across diverse biological systems. Understanding these dynamics can help in the design of more stable, synthetic microbiota that can reduce disease and improve fitness for a range of host species.
- Link to the paper in external page "Nature Microbiology"
- Link to external page "NCCR Microbiomes"