03
Julia Vorholt receives the Feodor Lynen Medal

Julia Vorholt, Professor at the Institute of Microbiology, Department of Biology, has been awarded the Feodor Lynen Medal 2024 for her work in the field of biochemistry. Congratulations!
DIP-MS, a deep interaction proteomics method for the analysis of protein complexes
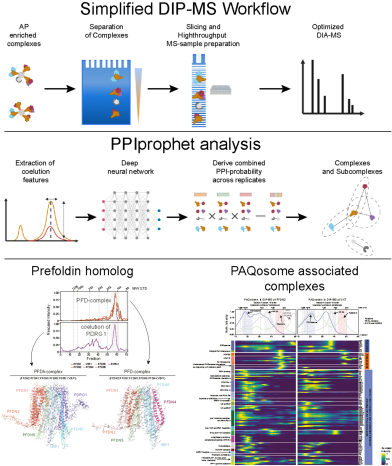
Deep Interactome Profiling (DIP-MS) offers a sensitive and comprehensive method to study protein complex isoforms concurrently present in affinity purifications. The method combines protein fractionation profiling with machine learning to study modular protein organisation at a high level of detail. The implemented method was developed in the Gstaiger and Aebersold lab (all IMSB) and has been recently published in “Nature Methods”.
Structure of antiviral drug bulevirtide bound to hepatitis B and D virus receptor protein NTCP
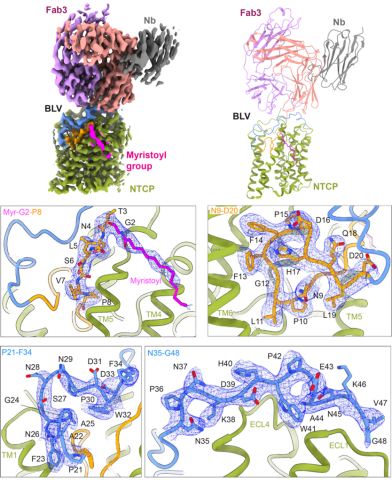
A recent article in the journal “Nature Communications” by the Locher group (IMBB) in collaboration with the Kossiakoff group (The University of Chicago), the Urban group (Heidelberg University), the Glebe group (Justus Liebig University Giessen), and the Geyer group (Justus Liebig University Giessen) describes the structural basis of mimicked HBV/HDV viral peptide drug interaction with its receptor NTCP.
FUS Droplets: Growing a Shell with Age

In a "Nature Chemical Biology" paper, the Allain lab (IBC) and collaborators including the Michaels group (IBC) mechanistically describe how droplets of the protein fused in sarcoma (FUS) mature. They grow over time a hard shell, which plays a critical role in droplet function.
Assessing microbiome population dynamics using genomic WISH-tags

To study the dynamics of microbiome assembly with strain-level resolution, the Vorholt, Hardt, and Sunagawa labs (all IMB) jointly developed a novel genomic barcoding system, the WISH-tags. These were used to assess how the order of arrival order impacts colonization in plant and mammalian microbiomes.