Systematic identification of structure-specific protein–protein interactions
A paper from the Picotti group (IMSB, in collaboration with the Beltrao, Riek and Korkhov groups) describes a method to screen for conformation-specific protein-protein interactions and pinpoint interaction sites. They used it to study interactors of the monomeric and amyloid states of alpha-synuclein.
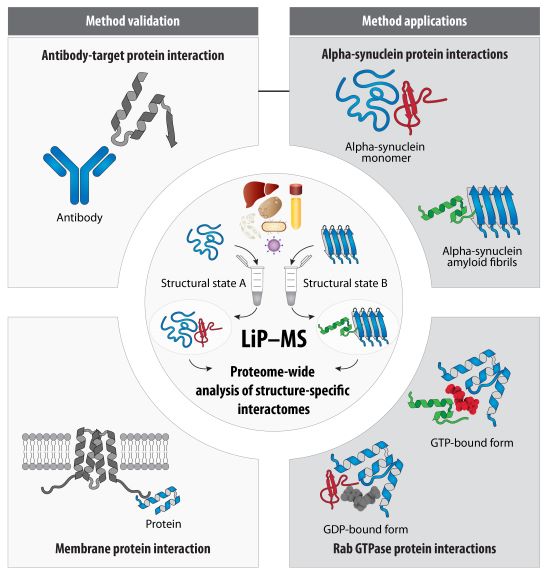
Proteins do not act alone but often function as protein complexes, in which they interact with other proteins in the cell. Further, many proteins can change their conformation, or shape, which may contribute to disease. For instance, changes in the protein alpha-synuclein from a small floppy monomer to a rigid multimeric fibril are suspected to be linked to Parkinson’s disease. In such a case, knowing the interactors of each structure may reveal molecular and cellular details of the disease.
There are many methods to identify interactors of a protein, but they are not practical to search for conformation-specific interactors. The Picotti group has now shown that limited proteolysis linked to mass spectrometry (LiP-MS) can be used for this purpose. They use a spike-in strategy, in which a bait protein in a structural state of interest is added to a lysate.
LiP-MS identifies changes in protease accessibility of proteins in two different conditions, such as with and without bait spike-in. The group showed, using known antibody-antigen interactions, that LiP-MS can identify protein-protein interactions in vitro and in lysate, and that it pinpoints known interaction sites. They identified known interactions of calmodulin with the membrane protein adenylyl cyclase 8. They also detected known interactors of alpha-Synuclein monomer and fibrils and report several novel structure-specific candidate interactors for this protein. Finally, they report candidate interactors of GTP- and GDP-bound forms of two Rab GTPases.
The approach will help elucidate structure-specific interactomes for any protein that can be purified in one or more stable structural states.
Link to the paper in external page "Molecular Systems Biology".