FilamentID reveals the composition and function of metabolic enzyme polymers
A recent "Cell" paper by the Pilhofer Lab (IMBB) and the Matos Lab (formerly IBC, now Max Perutz Labs in Vienna) unveils a multiscale filament identification workflow to characterize protein filaments that assemble during the developmental program of gamete formation in budding yeast.
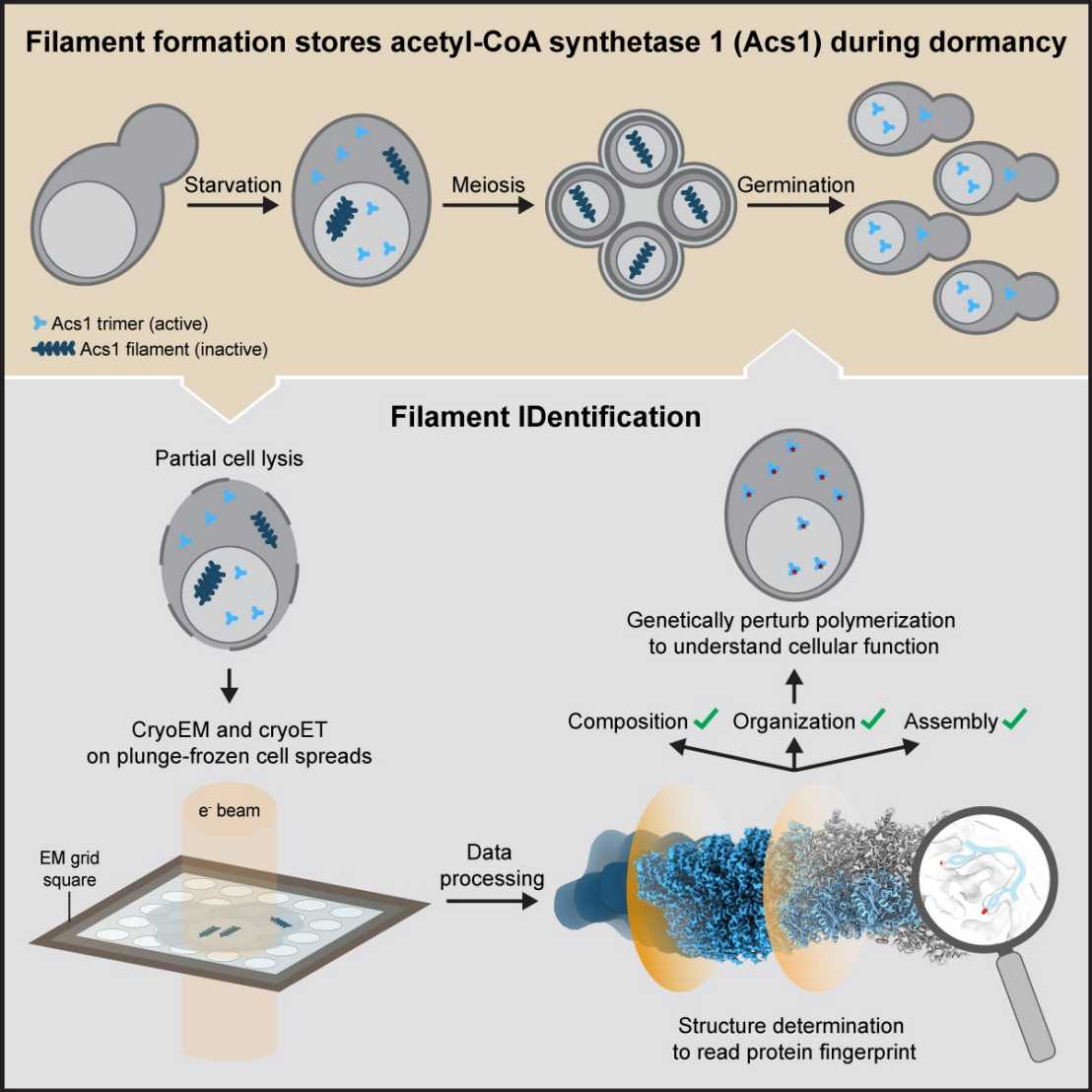
The process of gamete formation and subsequent development of progeny often involves extended phases of suspended cellular development or even dormancy. How cells adapt to endure such periods and subsequently return to growth remains poorly understood.
In their recent study, researchers visualized budding yeast cells undergoing gametogenesis by cryo-electron tomography. They discovered different types of elaborate filamentous assemblies within the nucleus, cytoplasm, and mitochondria.
To determine the composition of the filaments, the researchers developed a workflow to directly visualize partially lysed cells or organelles using a combination of cryo-electron microscopy imaging modalities. The “Filament Identification” (FilamentID) workflow revealed two conserved metabolic enzymes that assemble filament bundles. The aldehyde dehydrogenase Ald4 forms filaments in the mitochondria, whereas the acetyl-CoA synthetase Acs1 polymerizes into filaments in the nucleus and cytoplasm, respectively. Structural characterization provided molecular insights underlying polymerization and allowed for the targeted perturbation of filament assembly. Subsequent functional assays suggested that Acs1 filament formation facilitates the recovery of chronologically aged spores and, more generally, the cell cycle re-entry of starved cells.
The FilamentID workflow has the potential to be applied to investigate various filamentous macromolecular assemblies of unknown identity in diverse cellular contexts.
Link to the paper in external page "Cell" and an accompanying paper in external page “Developmental Cell”.