Non-canonical start codons enhance carbohydrate use in commensal E. coli within the mouse gut
A recent paper in "Nature Microbiology" by the Hardt group (ETH) and the von Mering group (UZH) uncovers a surprising role for non-ATG start codons in boosting lactose utilization in the gut, hinting at a wider impact on nutrient competition and probiotic efficacy.
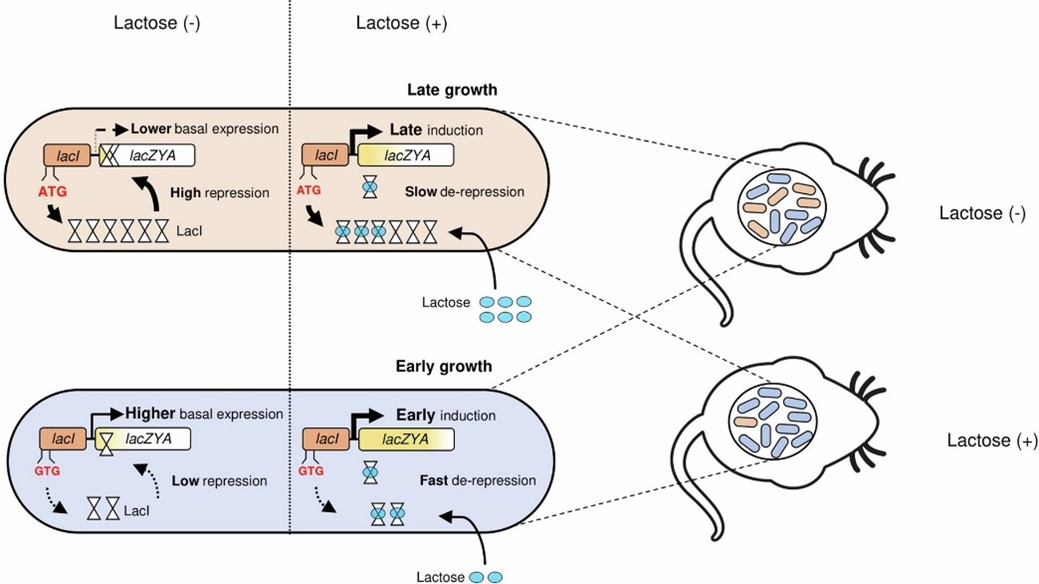
Researchers have discovered a surprising way that gut bacteria can outcompete their rivals, which could have important implications for our understanding of the gut microbiome composition and stability. Typically, bacteria compete by using up essential nutrients that others need to grow. However, this new study looked at how two nearly identical bacteria could still have different success rates when competing for the same nutrient—in this case, lactose.
The research focused on Escherichia coli (E. coli) 8178, a common gut bacterium in mice. Scientists found that a small genetic difference in how the bacteria start producing a specific protein, which helps them process lactose, gives one strain an edge over the other. This difference comes down to a non-standard "start" signal in the bacterium's DNA, called a non-ATG start codon, which subtly increases the bacterium's ability to break down lactose.
In experiments, the E. coli strain with this non-standard start codon outcompeted a similar strain with the more common start codon, especially when lactose was readily available. However, when lactose levels were reduced, the advantage was mitigated, showing that the benefits of this genetic tweak depend on the environment.
This discovery not only sheds light on how tiny genetic changes can impact bacterial success in the gut but also suggests that non-standard start codon might play a broader role in how bacteria compete for resources. This could have future implications for developing probiotics and understanding how gut bacteria maintain their balance.
Link to the paper in external page "Nature Microbiology".