Removal of TREX1 activity enhances CRISPR–Cas9-mediated homologous recombination
A recent “Nature Biotechnology” paper by the Corn group (IMHS) in collaboration with Maciejowski group from Memorial Sloan Kettering Cancer Center identified a novel molecular factor suppressing CRISPR-Cas homology-directed repair.
Have you ever wondered why CRISPR-Cas mediated HDR editing is so efficient in some cells but terribly inefficient in others? Struggling with gene editing in your cells? We've got the solution you've been looking for!
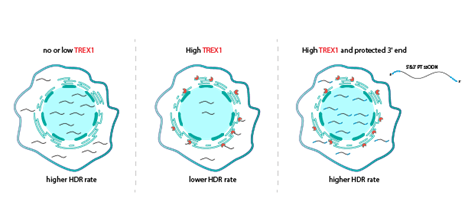
We embarked on a journey to identify methods for enhancing gene editing efficiency in Fanconi anemia (FA) cells, which harbor mutations in FA proteins necessary for optimal HDR efficiency. Our genome-wide CRISPRi screen in FA patient-derived cells revealed a single critical target: TREX1. TREX1 plays a critical role in reducing HDR efficiency when the repair template is either single-stranded or linearized double-stranded DNA. TREX1 expression serves as a biomarker for CRISPR-Cas9-mediated HDR, and high levels of TREX1, observed in various cell types including U2OS, Jurkat, MDA-MB-231, primary T cells, and hematopoietic stem and progenitor cells (HSPCs), are predictive of poor HDR outcomes. Moreover, we have demonstrated that HDR efficiency can be significantly improved through either knockout of TREX1 or the use of chemically protected single-stranded DNA templates that are resistant to TREX1 activity. Namely, phosphorothioate 3’ end protection is sufficient for fast inexpensive improvements to HDR in contexts with appreciable TREX1 expression. These strategies offer promising avenues for enhancing CRISPR-Cas9–mediated HDR, particularly in cell types with high TREX1 expression.
Link to the paper in external page "Nature Biotechnology".