The hGID GID4 E3 ubiquitin ligase complex targets ARHGAP11A to regulate cell migration
A recent "Life Science Alliance" paper by the Peter group (IBC), in collaboration with the Côté (IRCM, Montréal, Canada) and Pertz groups (ICB, Bern), demonstrates a novel role for the human GID E3 ligase in regulating cell migration by targeting the RhoA-GAP ARHGAP11A for proteasomal degradation.
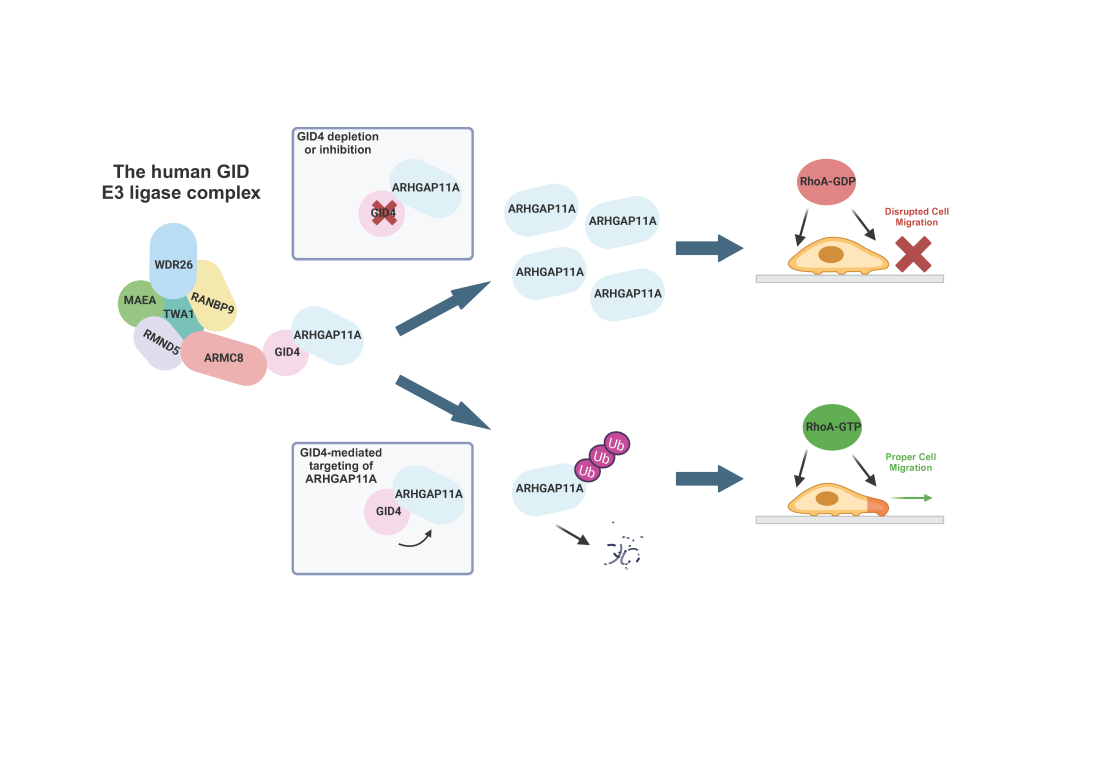
The CTLH/GID complex is a conserved E3 ubiquitin ligase that in yeast regulates glucose metabolism by degrading gluconeogenetic enzymes with an N-terminal degron motif. However, the function and regulation of the human CTLH/GID complex (hGID) remains largely unknown.
To address this, Dr. Bagci and colleagues deployed proximity-dependent biotinylation (BioID2) to identify proteins interacting with the hGID complex, revealing candidate substrates that specifically interact with its substrate receptor GID4. Among these, the RhoA-GAP ARHGAP11A was characterized as a key target, and hGIDGID4-dependent ubiquitination promotes its subsequent degradation via the proteasome. Surprisingly, ARHGAP11A lacks an N-terminal degron, suggesting that hGID4 may use distinct binding motifs. Notably, inhibition of GID4, either through genetic depletion or blockage of its substrate binding pocket using the PFI-7 inhibitor, leads to an accumulation of ARHGAP11A at the cell periphery. Interestingly, this stabilization inactivates RhoA, inhibiting cell motility and directional movement. Overall, these findings broaden the known substrate repertoire of the hGIDGID4 E3 ligase and reveal its critical role in regulating cell migration by modulating ARHGAP11A levels.
Link to the paper in external page "Life Science Alliance".