A rewritten bacterial genome to reveal control elements and genome design principles
Error-free and robust programming of biological systems necessitates rules to describe genome design. The Christen Group compares transcription of native and rewritten genes at the genome scale and contributes to genome annotation and the formulation of design principles in “Nature Communications”.
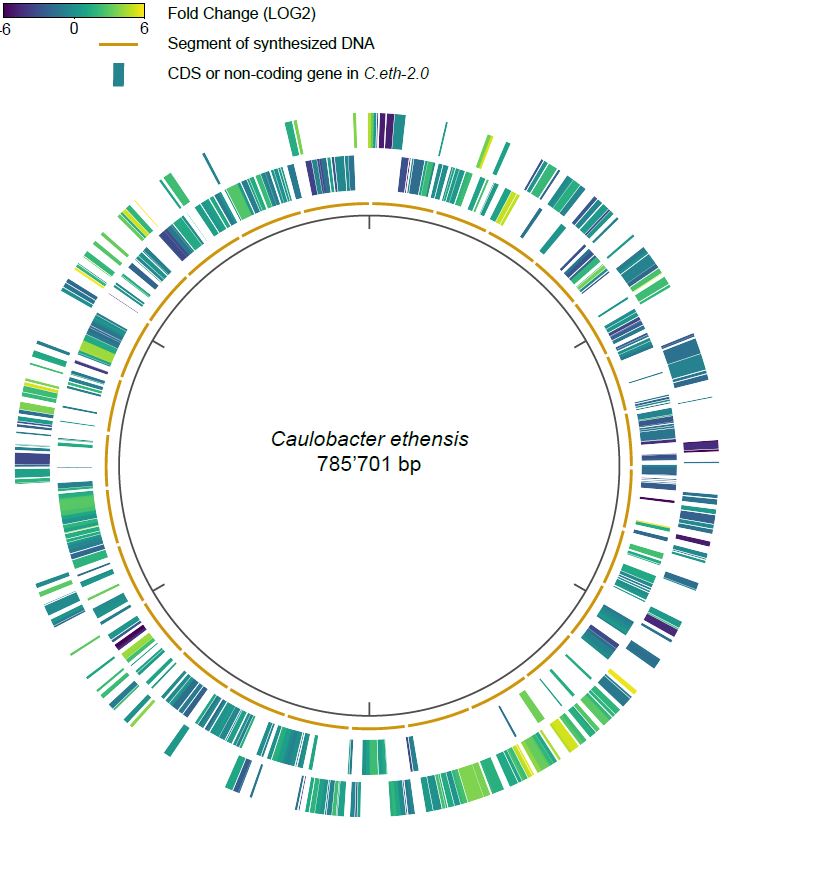
Native nucleotide sequences can be rewritten to enable DNA synthesis and assembly at scale and to create orthogonal genetic codes. Rewriting can be detrimental when sequences are interpreted in an unexpected way in the cell. In a systematic effort to better understand the underlying processes, the Christen Group has measured how a cell interprets genes in a rewritten bacterial genome at the level of the RNA, the messenger molecules that all cells make based on their DNA sequences.
The authors build on previous work, see https://ethz.ch/en/news-and-events/eth-news/news/2019/03/bacterial-genome-created-with-computer.html, in which they reported to have designed, synthesized and assembled a bacterial genome based on the genome of freshwater bacterium Caulobacter crescentus.
Christen and coworkers hypothesized that the changes that were introduced in the genome would lead to differences in the interpretation of the native and the rewritten genes. These changes would thus help to better understand how bacteria encode information in their DNA. As an example, the researchers uncovered an element in the RNA that helps to translate a ribosomal protein. Ribosomes are the universal machinery that translate RNA molecules to proteins in our cells. Based on instances in which the interpretation of rewritten DNA shows adverse effects on gene expression, the authors describe 4 design principles that form a contribution to a growing set of rules in the field of synthetic biology. The ultimate goal is to gather enough rules to describe very precisely how to program biological systems, with unlimited potential in bio- and health technology, and allowing to explore possibilities that evolution has not yet touched upon.
Link to the paper in external page Nature Communications