METALIC reveals interorganelle lipid flux in live cells by enzymatic mass tagging
A recent study in "Nature Cell Biology" by the Peter group (IBC) and the former Kornmann group (now at the University of Oxford), demonstrates a new versatile strategy to track the movement of lipids between organelles in vivo, using a combination of enzyme-based chemical modification and mass spectrometry.
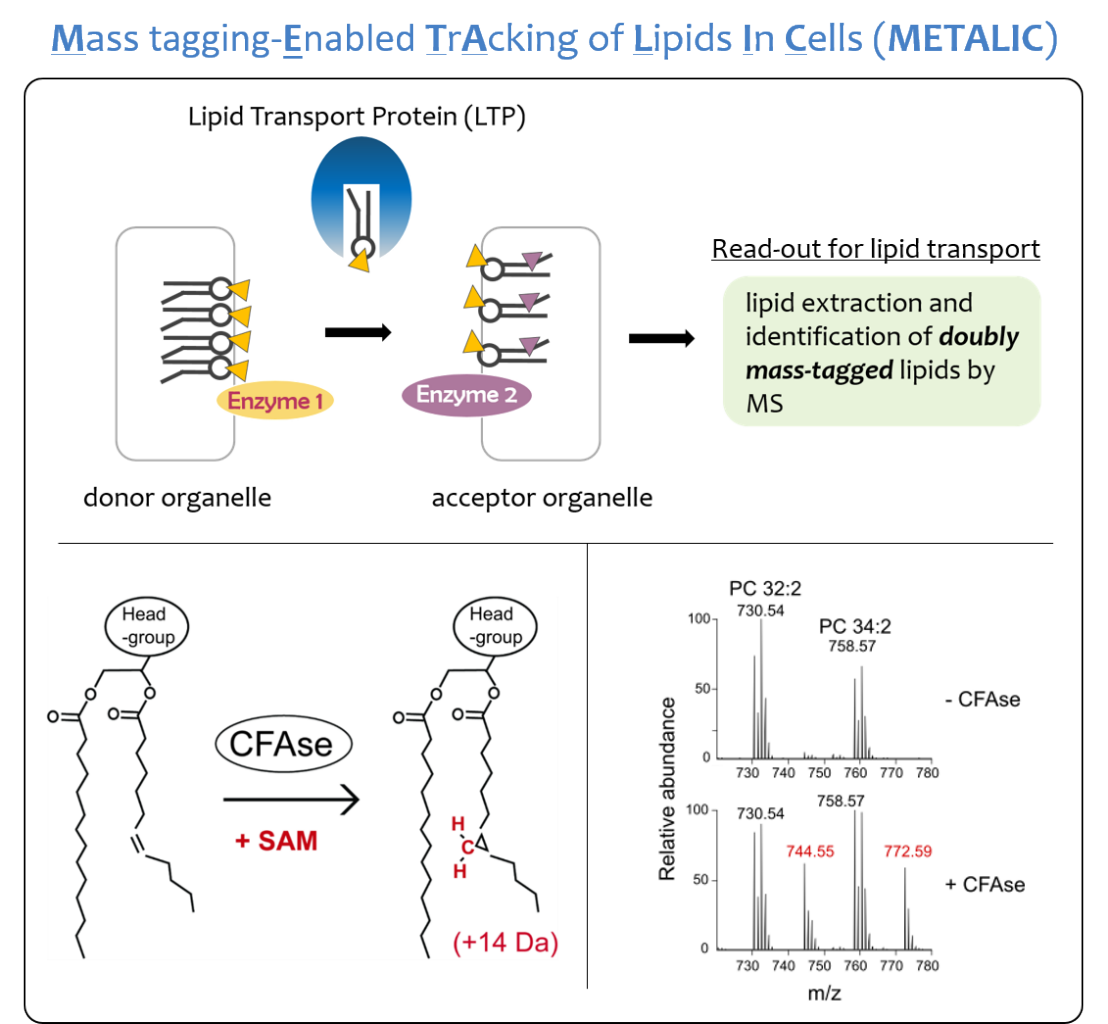
The distinct activities of organelles depend on the unique set of structurally diverse lipids that constitute their membranes. As lipid biosynthesis in eukaryotic cells mostly happens in the endoplasmic reticulum (ER), lipids must be transported to all other cellular membranes. Therefore, a major quest in cell biology has been to understand how lipids are transported between organelles. Several lipid transport proteins (LTPs) have been proposed to participate in lipid distribution based on in vitro assays, but despite their basic importance, in vivo evidence linking the absence of putative transport pathways to specific transport defects remains scarce. A reason for this scarcity is the near absence of in vivo lipid trafficking assays.
Here we introduce a versatile method named METALIC (Mass tagging-Enabled TrAcking of Lipids In Cells) to track interorganelle lipid flux inside cells. In this strategy, two enzymes, one directed to a ‘donor’ and the other to an ‘acceptor’ organelle, add two distinct mass tags to lipids. Mass-spectrometry-based detection of lipids bearing the two mass tags is then used to quantify exchange between the two organelles. Using METALIC, we show that two protein complexes, namely ERMES and Vps13–Mcp1, implicated in lipid transport have transport activity in vivo, and unravel their relative contributions to ER–mitochondria lipid exchange.
Despite the central contribution of lipids to a myriad of cellular functions, our knowledge on them lags behind DNA, RNA and proteins, as we do not have the equivalent tools (PCR and green fluorescent protein tagging). The development of METALIC thus takes an important step forward and paves the way to elucidate LTP function and lipid transport processes in vivo.
Link to the paper in "external page Nature Cell Biology"