Epigenetic and gene expression novelty in polyploids largely reverts during evolution
A recent study from the Bomblies group (IMPB) in Molecular Biology & Evolution investigates chromatin accessibility and transcriptomic changes upon genome duplication as well as subsequent evolution in Arabidopsis arenosa, showing immediate responses to polyploidy can revert during evolution.
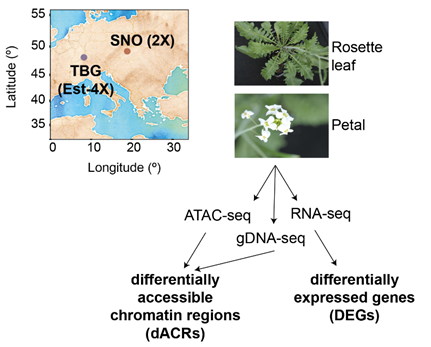
Whole-genome duplication (WGD), resulting in polyploidy, has been important in the evolution of all eukaryotes, but is particularly common in plants. Newly formed polyploids often show beneficial traits such as increased fruit size and stress resilience, but not all traits persist in evolved polyploids. Does novelty connect to stochastic epigenetic or transcriptomic changes? Do these persist over evolutionary time?
Srikant et al. address this using the plant Arabidopsis arenosa, which naturally occurs in diploid and autotetraploid forms allowing study of longer-term consequences of WGD, while new tetraploids can be made in the lab from diploids to study immediate consequences of WGD. The authors used ATAC-seq (a sequencing approach to identify accessible chromatin regions) and RNA-seq (to measure gene expression) on leaf and petal samples from diploid, newly-tetraploid, and evolved tetraploid A. arenosa. >8000 genomic regions differ in accessibility between the three, with patterns largely tissue specific. ~70% arise upon WGD, but most return to “diploid-like” in evolved tetraploids. Interestingly, only a small proportion of differentially accessible regions had associated changes in nearby gene expression, suggesting they may have a more structural role. Variation in accessibility correlated with nearby transposable elements. Relatively few genes were differentially expressed upon genome duplication, and ∼60% of these reverted to near-diploid levels in evolved tetraploids, again suggesting most initial perturbations do not last.
Link to the paper in external page "Molecular Biology and Evolution".