Pareto optimality couples metabolic noise to phenotypic heterogeneity in Escherichia coli
Heterogeneity between genetically identical bacterial cells can have important functional consequences, like increased survival upon antibiotic treatment. A collaborative effort between the Zampieri (IMSB) and Jenal (Biozentrum, University of Basel) groups funded by the NCCR AntiResist unravels the role of metabolism in mediating the propagation of noise in gene expression to phenotypic heterogeneity.
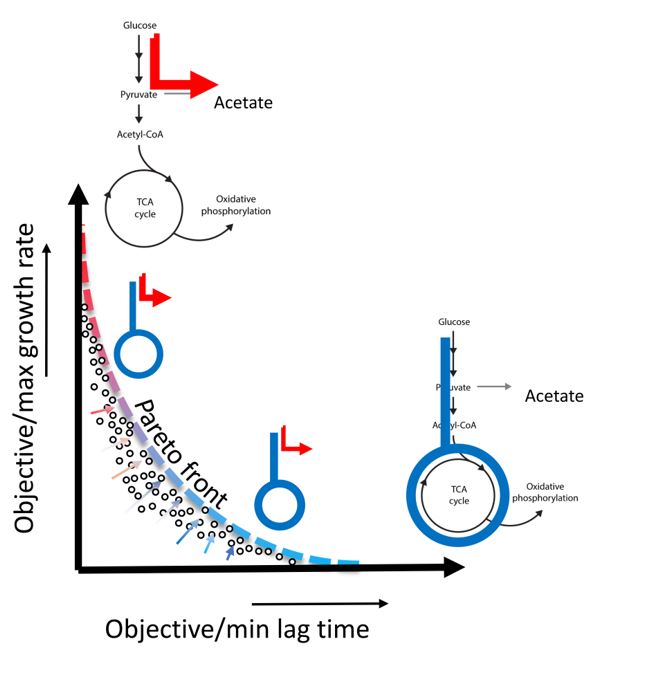
The nature of the evolutionary forces that shape cellular noise and the mechanisms translating such noise into phenotypic heterogeneity have remained elusive. The research teams at ETH and Biozentrum discovered unexpectedly high levels of noise in key enzymes of the central metabolism of E. coli, implying a large diversity of respiro-/fermentative metabolism in isogenic bacterial populations. To unravel the role of metabolic diversity in mediating phenotypic heterogeneity, the authors developed a multiparametric assay that monitors phenotypic and metabolic heterogeneity of E. coli growing on solid agar media. This approach allowed them to investigate the relationship between variations in respiratory activity and phenotypic parameters like maximum growth rate and lag time. The authors found that these variations follow Pareto optimal tradeoffs between maximization of growth rate and minimization of lag time, two objectives competing between fermentative and respiratory metabolisms. Based on this, they propose that Pareto optimality plays a key role in shaping and preserving phenotypic heterogeneity and in allowing bacteria to adapt to fluctuating environments.
Link to the paper in external page Nature Communications.
Comments
No comments yet