Single-cell landscape of innate and acquired drug resistance in acute myeloid leukemia
In a recent publication in Nature Communications, researchers from the Tumor Profiler consortium combined single-cell multi-omics and functional profiling for 21 AML patients to reveal resistance mechanisms to the Bcl-2 inhibitor venetoclax and identify emerging treatment vulnerabilities.
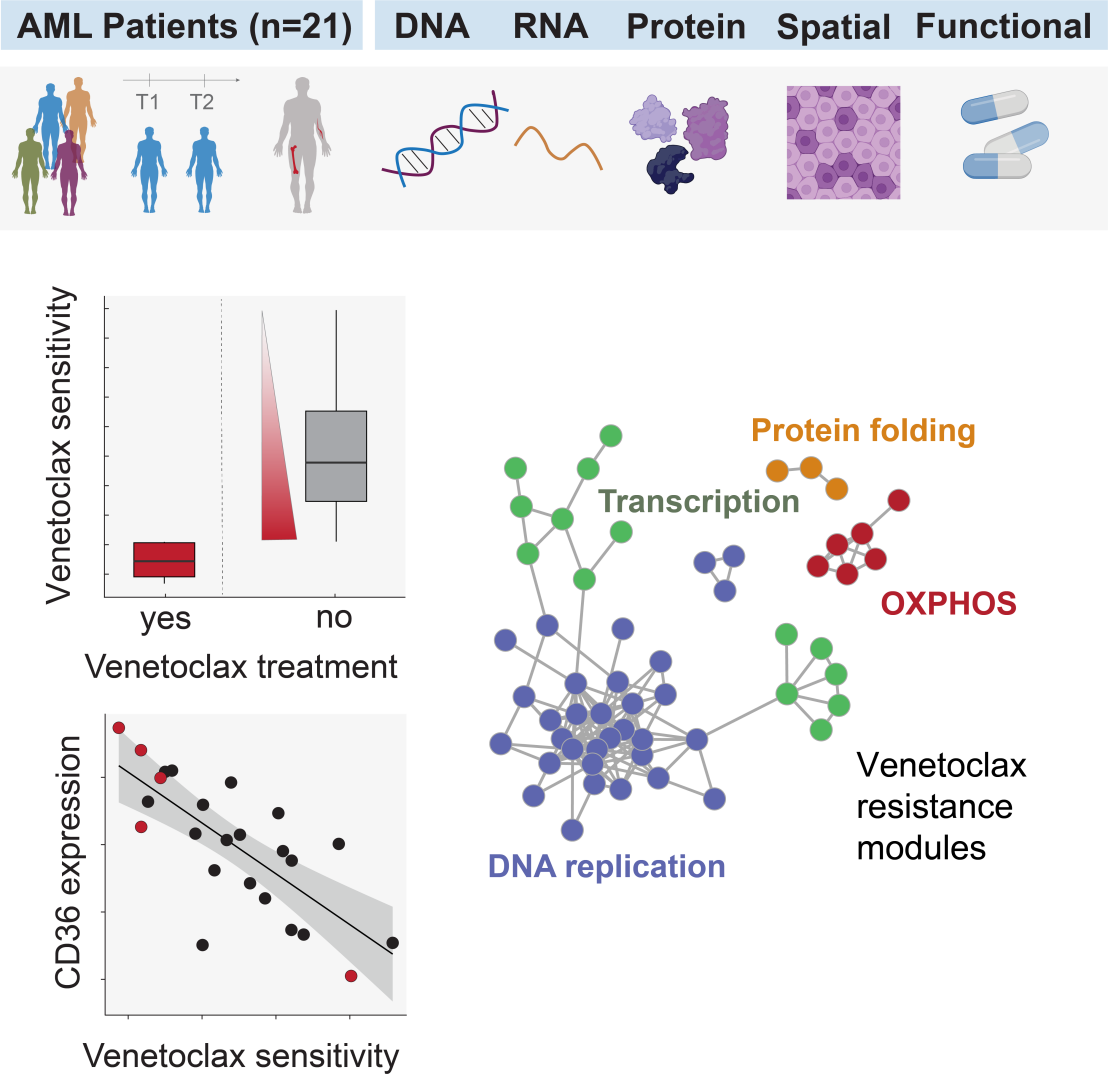
Molecular and functional profiling technologies (“Multi-OMICS”) open new doors for personalized medicine. The Tumor Profiler study (external page Irmisch et al, Cancer Cell, 2021) is a collaboration between Swiss universities, hospitals and industry with the goal to assess the clinical relevance of in-depth tumor profiling. In a recent publication of the acute myeloid leukemia (AML) arm of the Tumor Profiler, researchers demonstrated that multi-OMICS profiles can be generated in a clinically meaningful turnaround time and are highly informative of treatment resistance and sensitivity. In this study, they analyzed samples from 21 AML patients and combined up to nine OMICS methods to examine cellular drug responses and individual cell DNA, RNA, and proteins. They found that prior venetoclax exposure was associated with weaker response to this drug and accompanied by characteristic molecular changes. These changes were also observed in some venetoclax-naïve patients whose cells nevertheless did not respond to this treatment ex vivo. This analysis uncovered venetoclax resistance mechanisms and pointed towards potential alternative treatments targeting the resistance-associated molecular pathways. For instance, high levels of the protein CD36 in resistant cells were linked to ex vivo responses to a new immunotherapy. This study underscores the power of advanced profiling techniques in uncovering drug resistance mechanisms and identifying alternative treatment options for AML patients.
The AML arm of the Tumor Profiler study was co-led by Alexandre Theocharides (USZ), Berend Snijder (ETHZ), and Markus Manz (USZ), as well as Bernd Bodenmiller, Bernd Wollscheid, Lucas Pelkmans, Gunnar Rätsch, Mitch Levesque, Viola Heinzelmann-Schwarz, Ruedi Aebersold, Viktor H. Koelzer, Marina Bacac, Andreas Wicki, Holger Moch, Niko Beerenwinkel and includes over 100 Tumor Profiler consortium members. The study received funding from the PHRT - Personalized Health and Related Technologies, Roche, ETH Zürich, University of Zurich, University Hospital Zurich, and University Hospital Basel.
Paper (open access):external page external page https://doi.org/10.1038/s41467-024-53535-4
Tumor Profiler consortium: external page https://tumorprofilercenter.ch/
Comments
No comments yet